Published online Feb 26, 2018. doi: 10.13105/wjma.v6.i1.1
Peer-review started: December 4, 2017
First decision: December 27, 2017
Revised: January 17, 2018
Accepted: February 24, 2018
Article in press: February 25, 2018
Published online: February 26, 2018
Processing time: 93 Days and 13.8 Hours
To evaluate the most common studied genetic polymorphisms that may have an etiological role in irritable bowel syndrome (IBS).
The data base PubMed was searched for studies analyzing the association between gene polymorphisms and IBS. All original full papers, written in English, were retained for further analysis. The retrieved papers were further systematized according to those polymorphisms that have been detected in IBS.
Considering these criteria, our literature search found 12 polymorphisms, residing in 10 genes, which were reported to be consistently associated with IBS. The initial search identified 189 articles, out of which 48 potentially appropriate articles were reviewed. Of these 48 articles, 41 articles were included in the review. These articles were published between 2002 and 2016. Out of these 41 studies, 17 reported analysis of the serotonin transporter (SERT) gene (SLC6A4), eight on guanine nucleotide-binding protein subunit beta-3 (GNbeta3), six on the serotonin type 3 receptor genes (HTR3A), four on (HTR3E), three on (HTR2A), three the tumor necrosis factor superfamily member TL1A gene (TNFSF15), and ten on genetic polymorphisms with limited evidence.
Current evidence for the relation between genetic polymorphisms and IBS is limited owing to the fact that high-quality prospective studies and detailed phenotyping of patients suffering from IBS and matched controls were lacking in the past.
Core tip: The main genetic polymorphisms encountered in irritable bowel syndrome (IBS) are: Serotonin transporter (SERT) gene (SLC6A4), guanine nucleotide-binding protein subunit beta-3 (GNbeta3), serotonin type 3 receptor genes (HTR3A), (HTR3E), (HTR2A), the tumor necrosis factor superfamily member TL1A gene (TNFSF15). We performed a review of existent data, that studied genetic polymorphisms in IBS patients. We found that the actual IBS subgroups are not sufficient in order to identify distinct phenotypes and further in leading to new guiding principles for treatment. This systematic review demonstrates the need for genetic studies with an increasing number of subjects, because contradictory findings in terms of IBS subtype have been reported.
- Citation: Popa SL, Dumitrascu DL, Vulturar R, Niesler B. Genetic studies in irritable bowel syndrome-status quo. World J Meta-Anal 2018; 6(1): 1-8
- URL: https://www.wjgnet.com/2308-3840/full/v6/i1/1.htm
- DOI: https://dx.doi.org/10.13105/wjma.v6.i1.1
Irritable bowel syndrome (IBS) is the main digestive functional disorder, with a prevalence of 10%-20% of the population and has multifactorial etiology since genetic predisposition and environmental factors shape the phenotype.
According to the Rome IV criteria, the syndrome is defined as recurrent abdominal pain on average at least 1 d/wk in the last 3 mo, associated with two or more of the following symptoms: related to defecation, associated with a change in the frequency of stool, associated with a change in form (consistency) of stool. Classifying patients with IBS into specific subtypes based on predominant bowel habits is useful because is focusing the treatment on the predominant symptom. Accordingly to the Rome IV Criteria, IBS is classified into four subtypes: IBS with predominant constipation (IBS-C), IBS with predominant diarrhea (IBS-D), with mixed bowel habits (IBS-M) or unsubtyped (IBS-U). Patients meet diagnostic criteria for IBS-U if their bowel habits cannot be accurately categorized in any of the above subtypes[1]. The genetic predisposition is underlying the pathogenesis and the pathophysiology of IBS. Studies that point out higher concordance rates of monozygotic twins compared to dizygotic twins suggest that there may be distinct molecular bases for all IBS subtypes and genes that control neuronal function, the epithelial barrier integrity, mucosal immune interactions with bacteria in the gut. Unfortunately, the number of studies about single nucleotide polymorphisms (SNP) in selected candidate genes associated with IBS is still small.
The aim of this study was to review the existing literature on genetic polymorphisms associated with IBS.
A PubMed search was carried out in September 2016, looking for published papers analyzing the association between gene polymorphisms and IBS. Search keywords were: IBS and gene polymorphism. The inclusion criteria were: original articles that included patients with IBS-C, IBS-D or IBS-M, and that studied genetic polymorphisms in IBS patients. Exclusion criteria were: reviews, lack of abstract, non-English publications. Furthermore, ethical background was taken into account. We decided not to analyze SNP, which is less investigated, we only found it reported in five papers, because we decided that they are not relevant and may introduce bias.
As a result of our literature survey, we were able to review 12 polymorphisms, residing in 10 genes. All of them are considered to be associated with IBS (Table 1).
| Gene | SNP | Polymorphism | IBS type | Diagnostic criteria | Number of articles | Ref. |
| SLC6A4 | rs4795541 | 5-HTTLPR (-1950- 1949insT, -1950-1949insC), STin2.9 VNTR | IBS-C | Rome I, II, III | 15 | [2-5,7-12,14,15,19,22,27] |
| rs25531 | 179A > G (-1936A > G) | Rome II, III | 4 | [8-10,17] | ||
| HTR2A | rs6311 | -1438G > A (-998G > A | IBS-D | Rome I, II, III | 1 | [4] |
| rs6313 | 102C > T | IBS-D | Rome I, II, III | 2 | [4,35] | |
| HTR3A | rs1062613 | 42C > T; 178C > T (-24C > T) | IBS-D | Rome I, II, III | 6 | [2,16,20-22,45] |
| HTR3E | rs56109847 | 76G > A | IBS-D | Rome I, II, III | 5 | [16,18,20,21,45] |
| GNB3 | rs5443 | 825C > T | IBS-C | Rome II, III | 8 | [23-25,29,32,34,35] |
| TNFSF15 | rs4263839 | A/G | IBS-C | Rome I, II, III | 3 | [6,26,28] |
| Limited number of studies: pV158M CCK rec.intron1 NXPH1CDC42 | Rome III | 10 | [30,31,33,36-40,43,44] |
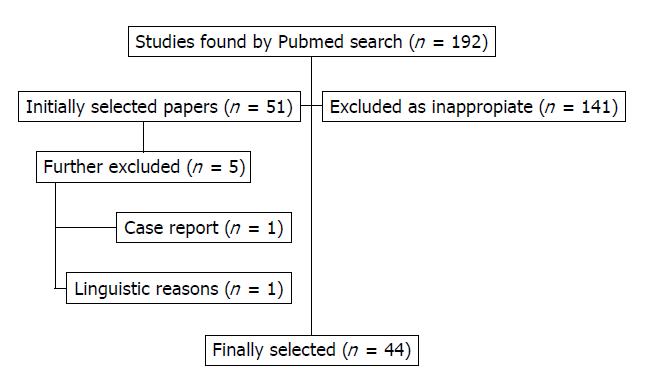
The initial search identified 182 articles, out of which 48 potentially appropriate articles were reviewed. Of these 48 articles, 44 articles were included in the review. These articles were published between 2002 and 2016. Out of these 44 studies, 20 reported analysis of the serotonin transporter (SERT) gene (SLC6A4), eight on guanine nucleotide-binding protein subunit beta-3 (GNbeta3), six on the serotonin type 3 receptor gene (HTR3A), four on (HTR3E), three on (HTR2A), three the tumor necrosis factor superfamily member TL1A gene (TNFSF15), and ten on genetic polymorphisms with limited evidence (Figure 1).
In the following we will describe the reported evidence of a relationship between gene polymorphisms and IBS published to date.
Serotonin (5-hydroxtryptamine, 5-HT) is an essential neurotransmitter involved in regulation of gut function, by playing key roles in intestinal peristalsis and in sensory functions mediated via the brain-gut axis. The serotonin transporter (SERT) encoded by the gene SLC6A4 regulates the intensity and duration of serotonin signaling by reuptaking serotonin from the synaptic cleft, thereby terminating its efficacy. This makes it an excellent candidate gene for analysis of genetic predisposition to IBS.
Disturbance in serotonin reuptake can modify enteric signaling, leading to gut dysfunctions, thereby contributing to the pathophysiology of IBS.
The solute carrier family 6 member 4 (SLC6A4) gene encodes the serotonin transporter (SERT). Polymorphisms in the promoter region of the SERT gene has a direct effect on transcriptional activity, which may result in altered 5-HT reuptake activity. The investigation of the association between 5HTTLPR in the SERT gene and IBS, using subgroup population-based analysis, point out that visceral hypersensitivity in IBS can be related to genetic factors[2-6]. To date, the S allele in the promoter region as well as the STin2.9 VNTR allele residing in an intron, have been reported to be related to anxiety and depression, a result that supports a biopsychosocial model of IBS, with the genotype in SLC6A4 that is increasing the risk for depressive episodes. Increased risk of IBS-C is presented by individuals with of L/L genotype and 12/12-L/L genotype association[5,6]. IBS-D and IBS-A are more frequent in individuals with L/S genotype[6]. Other studies suggest that the s/L polymorphism of serotonin transporter gene is linked only with the IBS-C development, this link being present only in East Asian population[7]. Moreover, the response to tegaserod was influenced by the genotype: L/L being poorer than S/S and S/L genotypes[5]. Carriers of S allele in 5-HTTLPR region was published as being frequent in Chinese Han population, with IBS, but other associations studies looking for IBS and variable number of tandem repeats (VNTRs) and tag SNPs, such as rs1042173, rs3794808, rs2020936 in SERT gene [using polymerase chain reaction (PCR) and TaqMan® SNP Genotyping, and positive haplotype], were not found[8]. SLC6A4-polymorphism and higher levels of 5-HT (in rectal biopsy of patients) were significantly linked with IBS-D and abdominal pain, suggesting that SLC6A4 has an important role in IBS pathophysiology[9,10]. Also in IBS-D, platelet SERT is reduced and is related with low levels of SERT mRNA.
A metaanalysis by Zhang et al[11] looked to 25 studies including more than 3000 patients with IBS and more than 3000 controls (diagnosed with different criteria according to the moment of the study: Rome I/II/III). The meta-analysis showed that the 5HTTLPR L allele and L/L are involved in the IBS-C development, in East Asian population, but not Central Asian populations.
On the contrary, other studies found a negative association between IBS and 5HTTLPR in the SERT gene. This is the case of a metaanalysis by Areeshi et al[12], which analyzed 12 studies with over 2000 IBS cases and over 2000 The same lack of association is found in the studies that have examined another SERT gene polymorphism, STin2 (located in intron 2), with undetermined ethnicity[13].
A study on a group of North American Caucasian female patients with IBS-D, analyzed leukocyte DNA, by polymerase chain reaction, for nine SERT polymorphisms. The result was that SERT-P S/S genotype was significant associated with IBS-D[14]. On the other hand, a study on American and Asian populations demonstrates that SLC6A4 (S/L) polymorphism is associated with reduced risk of IBS[15].
The activation of different brain regions during colorectal distension in subjects carrying the S allele of the SERT gene SLC6A4 promoter polymorphism 5-HTTLPR, suggests that individuals with a reduced level of SERT may more intensively respond to gut signals in emotion-regulating brain circuit. The amygdala region is more activated during a fearful face recognition paradigm in fMRI studies. This data demonstrates the relation between visceral pain and the individuals with a weak function of serotonin transporter[23]. A study using the Rome I criteria, in 54 Turkish IBS patients, showed a high incidence of the C/C genotype for 102T > C, A/A genotype for−1438G > A, HTR2A gene, rs6313 and IBS-D[3]. Similar results were found in a Greek study, showing that the frequencies of the SS genotype and S allele of the serotonin transporter polymorphism were significantly associated with IBS and the TT genotype and T allele frequencies of G protein β3 subunit showed also significant difference between the IBS patients and healthy controls[2].
Other plausible candidates of the serotonergic system represent 5HT3 receptors (5HT3Rs) mediating the effects of 5HT on intestinal functions during the postprandial period. A sequencing study of the HTR3 genes in IBS detected the 5’-UTR variant c.-42C > T of HTR3A (rs1062613) and 3’-UTR variant c. 76G > A in HTR3E (rs62625044). They found an association of SNPs in HTR3A and HTR3E in patients with IBS-D in a cohort from the United Kingdom; in particular the SNP in HTR3E was replicated in another cohort from Germany[2,16]. A recent study that investigated the relation between these SNPs in HTR3A and HTR3E and IBS-D in 500 IBS-D Chinese patients and 500 healthy control subjects replicated these findings. The PCR-RFLP method revealed a significant difference in the SNP frequency between the IBS-D patients and the healthy control subjects in the distribution of genotype and the minor allele of rs1062613 in HTR3A gene. Moreover, data about rs62625044 in HTR3E gene, evidenced a significant difference between the distribution of GA genotype and A allele, only in female patients[16].
A small sample size study of pacients with IBS showed that the carriers of the rare G allele of rs25531 had approximately threefold increased odds to present IBS than healthy controls . Onwards, the G-allele was more frequent in diarrhea-predominant subjects than in constipation-predominant or alternator subjects[17].
Recent studies demonstrated that a functional variant (rs56109847) in the 3′-untranslated regions (3′-UTR) of the serotonin receptor 3E (HTR3E) gene associated with IBS-D in British populations is also present in IBS-D in the Chinese females, emphasizing the role of miR-510 on 5-HT3E expression of colonic tissues in patients with gastrointestinal disorders. Moreover, the mechanism that underlies the association of HTR3E SNP rs56109847 with IBS-D is also described. The 5-HT3E rs56109847 could directly inhibit the binding of miR-510 to HTR3E 3′-UTR in HEK293 and HT-29 cells and confirmed that the SNP (rs56109847) of the non-coding region of HTR3E affected the binding of mircoRNA, thus affecting the permeability of the GI tract[18].
In contradiction with the analysed data, a study shows that there is no association between the genetic polymorphism in the SERT-P gene and IBS. The fact that SERT-P polymorphism has recently been associated with treatment response is a further proof that the genetic polymorphism in the SERT-P gene might have a pharmacogenetic role[19].
Another more recent study replicated these findings in patients with IBS-D from Yangzhou, Jiangsu province, showing a significant difference between patients and the controls in HTR3A (rs1062613) and the frequency of T allele was significantly higher in both female and male patients than that in the controls (P < 0.05). They performed polymerase chain reaction (PCR) amplification and restriction fragment length polymorphism (RFLP) technique on DNAs from 300 healthy subjects and 450 patients with IBS-D[20]. Of note, the SNPs rs1062613 in HTR3A has initially been associated with major depression and “harm avoidance”, an inherited trait associated with depression and anxiety, frequently encountered in IBS. In a study from 2011, this SNP has been correlated with the severity of IBS symptoms, anxiety and changes in amygdala activity[15]. Alosetron, a selective 5HT3R antagonist, beneficial in the management of symptoms like abdominal cramping, stool urgency and diarrhea in women with IBS-D was investigated in a pharmacogenetics study[21]. This revealed a greater efficacy of slowing down colonic transit as evidenced by the fact that L/L compared to L/S or S/S carriers benefitted from the treatment, by being high responders. This seems to be plausible based on the hypothesis that L/L carriers, who are supposed to present with increased SERT expression, and consequently 5HT reuptake, may present lower synaptic 5HT levels and therefore less competition between endogenous 5HT and alosetron[22].
Catechol-O-methyltransferase (COMT) is an enzyme that degrades dopamine, epinephrine, norepinephrine and the functional polymorphism pV158M has most extensively analyzed to date in various conditions. The Val alleles lead to four-fold higher enzymatic activity compared to the Met allele and thereby may influence metabolic levels of its substrates[23]. The gene variant has been demonstrated, to play an essential role in processes associated with abstract thought, task structure, and the placebo effect[23,24].
It is well established that depression, anxiety and pain syndromes are related to altered COMT activity, conditions showing also a high co-morbidity with IBS. Consequently it presented another plausible candidate to be explored in the context of IBS. In a recent study from Sweden, the V/V genotype had a significantly higher occurrence compared with controls, but V/M genotype, had a lower occurrence in IBS compared with controls and exhibited significantly increased bowel frequency[24]. In elderly Chinese patients (over the age of 60 years), COMT158Met was related with IBS and significantly more prevalent in patients with IBS-D. Furthermore, it was prevalent in those patients with symptomatology that persisted over 5 years[25].
Tumor necrosis factor superfamily-15 gene (TNFSF15, also known as VEGI or TL1A) is a cytokine that has main functions in angiogenesis, immune system mobilization and inflammation. TNFSF15 stimulates T cell activation, Th1 cytokine production, dendritic cell maturation and inhibits endothelial cell proliferation and endothelial progenitor cell differentiation. The risk allele of the SNP rs4263839 G in TNFSF15 was initially associated with an increased risk of IBS, more pronouncedly, IBS-C[6].
In respect to postinfectious IBS(PI-IBS) it has been hypothesized that polymorphisms in genes whose expression were altered by gastroenteritis might be linked to IBS with diarrhea (IBS-D) which closely resembles PI-IBS[25]. Han et al[25] established an IBS-D association with rs6478109 and rs6478108, which are in linkage disequilibrium with rs4263839. In fact, they found indeed IBS-D and PI-IBS patients to be associated with TNFSF15 and TNF α genetic polymorphisms which also predispose to Crohn’s disease suggesting a possible common underlying pathogenesis. In addition, both SNPs are associated with TNFSF15 expression in colorectal tissue[27]. Furthermore, Czogalla et al[2] recently confirmed a modest association (OR 1.24) in IBS-C in a meta-analysis combining own validation data with published data from the two previous studies.
TL1A-Death Receptor 3 has an essential role in production of interferon-c and interleukin-17 via proliferation and differentiation of T-helper 17, explaining patterns of immune response in host-microbiota interaction with commensal bacteria that contribute to IBS risk. As well, data shows its implication in other inflammatory disorders[26-28].
Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-3 (GNβ3) is a protein that is encoded by the gene GNB3[28]. The G-protein is an important factor in intracellular signal transduction, mediating functions of ion channels and protein kinases. The SNP-825C > T is leading to a modified signal transduction of functional impact: changes of sensory function or motility associated with FGID (functional gastrointestinal disorders)[29]. The association of this polymorphism with IBS has been demonstrated, and recent data shows that alteration of GNB3 825C > T CC type has a direct effect on gastrointestinal sensitivity and peristalsis[30]. A group of elderly Chinese IBS evaluated using the Geriatric Depression Scale, was not able to relate the GNB3-825C > T SNP with IBS[25,31]. However, the TC/TT genotypes are associated with lower sensations of gas and urgency in response to rectal distention after administration of clonidine[32]. In line with this, a study on a group from Korea evidenced that the GNB3 825C > TT allele is associated with IBS-C and studies analyzing patients from Greece, also confirmed that the TT genotype and GNB3 T allele have a significant association with IBS[32-34].
A study in which two large independent IBS cohorts were genotyped to assess genetic variability in immune, neuronal and barrier integrity genes, determined that the following SNPs associated independently: rs17837965-CDC42 with IBS-C (OR exploratory = 1.59 (1.05 to 1.76); OR validation = 1.76 (1.03 to 3.01)) and rs2349775-NXPH1 with IBS-D (OR exploratory = 1.28 (1.06 to 1.56); OR validation = 1.42 (1.08 to 1.88)). The study included 935 IBS patients, 639 controls and 384 single nucleotide polymorphisms (SNPs) covering 270 genes. Other three SNPs in immune-related genes (rs1464510-LPP, rs1881457-IL13, rs2104286-IL2RA), one SNP in a neuronal gene (rs2349775-NXPH1) and two SNPs in epithelial genes (rs245051-SLC26A2, rs17837965-CDC42) were weakly associated with IBS (P < 0.05)[34-45].
The present review identified articles, most of them prospective studies, on genetic polymorphisms in IBS pathogenesis or after therapy. The major pitfall is that patients were recruited based on a non-uniform symptom classification: Rome I, Rome II, or Rome III in the studies that were taken into account. Study limitations were represented by language barriers of some articles (which prevented access), and the low number of patients involved in most of the studies (underpowered); the main reason for excluding articles was the insufficient number of studies on a particular genetic polymorphism and articles written in non-English publications. Other limitations of the meta-analysis were the intricacy of ethnicities, and the difficulty of taking multiple genotypes testing into account. Above all that, statistical results were rarely corrected for multiplicity. As a result false positive associations may have been reported.
The polymorphisms of the Serotonin transporter (SERT or SLC6A4) gene are the most frequent genetic polymorphisms studied in IBS to date. Studies proved that the A allele of HTR3E was significantly higher in female IBS-D patients and there were no differences in either A allele or GA genotype between male patients. A possible reason for why there is no association to be found in male, can be explained by the effect of ovarian hormones on visceral sensitivity. A supposition which needs to be verified by future research.
A recent meta-analysis of immunogenetic case–control association studies in IBS confirmed a moderate association of rs4263839 in TNFSF15, and particularly with IBS-C. Control samples recruited by harmonized criteria are essential in order to overcome limitations like low statistical power and large heterogeneity for studies of IBS.
Because of the limited number of studies, further studies are needed for the following polymorphisms: Cholecystokinin (CCK) is a peptide hormone responsible for stimulating the digestion of fat and protein and is produced by I-cells in the mucosal epithelium of the small bowel. It has the effect of releasing digestive enzymes and bile from the pancreas and gallbladder and recent data evidenced that low densities of secretin and CCK cells in IBS-diarrhea patients can cause a functional pancreatic insufficiency and also inadequate gall emptying[27,28].
Polymorphism in CCK receptor intron 1 was associated with IBS-C and IBS-M in Korean population[37,38]. Also with limited evidence is the adhesion between dendrites and axons, that is promoted by a tight complex with alpha neurexins and neurexophilin-1 a protein encoded by the NXPH1 gene. Genetic variants in NXPH1 are associated with IBS-D[6]. Cell division control protein 42 homolog (CDC42) is a protein with an essential role in cell cycle regulation, including cell structure, migration, endocytosis and cell cycle progression. Genetic variants in CDC42 are associated with IBS-D[39].
The biopsychosocial model of illness and disease, as first described by Engel, reconcileed the dualistic concept that separated illness and disease and is a good way to explain the interaction between cultural factors, ethnicity, geographic region, types of food, endocrinological factors, immunological factors and genetic markers, which exist in patients with IBS. Recent studies analyzing individual coping strategies, cultural level, education level, religious beliefs about health and disease, demonstrated that a biopsychosocial conceptualization of the pathogenesis and clinical expression of IBS is mandatory. Further, somatic symptoms interact with the psychological status and promote each other, making the investigation of IBS more difficult[40].
A recent study that analysed 288 103 participants from 41 countries, showed that the global prevalence of IBS has a significant degree of heterogeneity that ranged from 1.1% in France and Iran to 35.5% in Mexico, with significant variance in regional prevalence rates, from 17.5% (95%CI: 16.9% to 18.2%) in Latin America, 9.6% (9.5% to 9.8%) in Asia, 7.1% (8.0% to 8.3%) in North America/Europe/Australia/New Zealand, to 5.8% (5.6% to 6.0%) in the Middle East and Africa[41].
A major pitfall in the current genetic studies in IBS is represented by the low number of subjects included in the majority of studies. Fortunately, the number of centers around the world that are collecting samples is growing. Nevertheless no unified genetics workflow existed. From the genetic perspective, the actual IBS subgroups are not sufficient in order to identify distinct phenotypes and further in leading to new guiding principles for treatment. These limitations can be overcome by international cooperation, like the GENIEUR network (Genes in Irritable Bowel Syndrome Research Network Europe, http://www.genieur.eu), who allows the contribution of specialists from many countries and the collecting of large samples of subjects[42] who are deeply phenotyped to allow genotype phenotype correlation and data mining approached[42]. Such studies allow also the standardization of investigative tools in the approach of IBS patients[43-45].
In conclusion, Current evidence for the relation between genetic polymorphisms and IBS is limited owing to the fact that high-quality prospective studies and detailed phenotyping of patients suffering from IBS and matched controls were lacking in the past. Studies on functional gastrointestinal disorders and genetic polymorphisms analyzing the same genetic variants in comparably characterized case control cohorts are also very limited. Furthermore, association of TNFSF15 genetic polymorphisms, which also predispose to Crohn’s disease, suggest a possible common underlying pathogenesis. However, for both polymorphisms contradictory findings in terms of IBS subtype have been reported underlining the necessity of more detailed phenotypic information for data stratification. To date, the s/l polymorphism in SLC6A4, represents the most frequently studied polymorphism and the HTR3E SNP has been replicated in four studies to date.
The irritable bowel syndrome (IBS) is a hot topic and the uncovering its genetic determination is very important.
Knowing the genetic link in the occurrence of IBS could offer the perspective to better know this condition and to improve its management.
In order to shed light on this topic, we carried out a systematic review of the data on main genetic polymorphisms described uptoday.
A PubMed search was carried out in September 2016, looking for studies analyzing the association between gene polymorphisms and IBS. Search keywords were: IBS and gene polymorphism. The inclusion criteria were: original articles that included patients with IBS-C, IBS-D or IBS-M, and that studied genetic polymorphisms in IBS patients. Exclusion criteria were: reviews, lack of abstract, non-English publications.
The result of our study was a review of 12 polymorphisms, residing in 10 genes reported to be associated with the pathogenesis and the pathophysiology of IBS. The main problem that remains to be solved in the current genetic studies analysing IBS is represented by the low number of subjects included in the majority of studies.
High-quality evidence for the relation between genetic polymorphisms and the IBS etiology is lacking, as a result of the insufficient number of high-quality prospective studies. Similar studies on functional gastrointestinal disorders and genetic polymorphisms are also very limited. The strength of articles, included in this review are the determination of each genetic polymorphism, using high efficiency techniques. The polymorphisms of the Serotonin transporter (SERT or SLC6A4) gene were the most frequent genetic polymorphisms studied in this pathology. Investigation of PI-IBS patients showed associations with TNFSF15 genetic polymorphisms which also predispose to Crohn’s disease suggesting a possible common underlying pathogenesis.
From the genetic perspective, the actual IBS subgroups are not sufficient in order to identify distinct phenotypes and further in leading to new guiding principles for treatment. These limitations can be overcome by international cooperation, like the GENIEUR network (Genes in Irritable Bowel Syndrome Research Network Europe), who allows the contribution of specialists from many countries and the collecting of large samples of subjects who are deeply phenoytped to allow genotype phenotype correlation and data mining approached. Such studies allow also the standardization of investigative tools in the approach of IBS patients.
This manuscript results in part from collaboration and network activities promoted under the frame of the international network GENIEUR (Genes in Irritable Bowel Syndrome Research Network Europe), which has been funded by the COST program [BM1106, (http://www.genieur.eu)] and is currently supported by the European Society of Neurogastroenterology and Motility (ESNM, http://www.esnm.eu).
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country of origin: Romania
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Chiarioni G, Mihai C, Salerno Soares RL, Tandon RK, Wittmann T, Yuan JY S- Editor: Cui LJ L- Editor: A E- Editor: Li RF
| 1. | Schmulson MJ, Drossman DA. What Is New in Rome IV. J Neurogastroenterol Motil. 2017;23:151-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 419] [Cited by in RCA: 469] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 2. | Czogalla B, Schmitteckert S, Houghton LA, Sayuk GS, Camilleri M, Olivo-Diaz A, Spiller R, Wouters MM, Boeckxstaens G, Bermejo JL. A meta-analysis of immunogenetic Case-Control Association Studies in irritable bowel syndrome. Neurogastroenterol Motil. 2015;27:717-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Fukudo S, Kanazawa M, Mizuno T, Hamaguchi T, Kano M, Watanabe S, Sagami Y, Shoji T, Endo Y, Hongo M. Impact of serotonin transporter gene polymorphism on brain activation by colorectal distention. Neuroimage. 2009;47:946-951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 4. | Pata C, Erdal E, Yazc K, Camdeviren H, Ozkaya M, Ulu O. Association of the -1438 G/A and 102 T/C polymorphism of the 5-Ht2A receptor gene with irritable bowel syndrome 5-Ht2A gene polymorphism in irritable bowel syndrome. J Clin Gastroenterol. 2004;38:561-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Li YY, Nie YQ, Xie J, Tan HZ, Zhou YJ, Wang H. Serotonin transporter gene polymorphisms in irritable bowel syndrome and their impact on tegaserod treatment. Zhonghua Neike Zazhi. 2006;45:552-555. [PubMed] |
| 6. | Zucchelli M, Camilleri M, Andreasson AN, Bresso F, Dlugosz A, Halfvarson J, Törkvist L, Schmidt PT, Karling P, Ohlsson B. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut. 2011;60:1671-1677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Saito YA. Genes and irritable bowel syndrome: is there a link? Curr Gastroenterol Rep. 2008;10:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 8. | Yuan J, Kang C, Wang M, Wang Q, Li P, Liu H, Hou Y, Su P, Yang F, Wei Y. Association study of serotonin transporter SLC6A4 gene with Chinese Han irritable bowel syndrome. PLoS One. 2014;9:e84414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 9. | Kumar S, Ranjan P, Mittal B, Ghoshal UC. Serotonin transporter gene (SLC6A4) polymorphism in patients with irritable bowel syndrome and healthy controls. J Gastrointestin Liver Dis. 2012;21:31-38. [PubMed] |
| 10. | Foley S, Garsed K, Singh G, Duroudier NP, Swan C, Hall IP, Zaitoun A, Bennett A, Marsden C, Holmes G. Impaired uptake of serotonin by platelets from patients with irritable bowel syndrome correlates with duodenal immune activation. Gastroenterology. 2011;140:1434-1443.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 11. | Zhang ZF, Duan ZJ, Wang LX, Yang D, Zhao G, Zhang L. The serotonin transporter gene polymorphism (5-HTTLPR) and irritable bowel syndrome: a meta-analysis of 25 studies. BMC Gastroenterol. 2014;14:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Areeshi MY, Haque S, Panda AK, Mandal RK. A serotonin transporter gene (SLC6A4) polymorphism is associated with reduced risk of irritable bowel syndrome in American and Asian population: a meta-analysis. PLoS One. 2013;8:e75567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Pata C, Erdal ME, Derici E, Yazar A, Kanik A, Ulu O. Serotonin transporter gene polymorphism in irritable bowel syndrome. Am J Gastroenterol. 2002;97:1780-1784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 105] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Yeo A, Boyd P, Lumsden S, Saunders T, Handley A, Stubbins M, Knaggs A, Asquith S, Taylor I, Bahari B. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut. 2004;53:1452-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 182] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 15. | Choi YJ, Hwang SW, Kim N, Park JH, Oh JC, Lee DH. Association Between SLC6A4 Serotonin Transporter Gene Lainked Polymorphic Region and ADRA2A -1291C> G and Irritable Bowel Syndrome in Korea. J Neurogastroenterol Motil. 2014;20:388-399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Gu QY, Zhang J, Feng YC, Dai GR, Du WP. Association of genetic polymorphisms in HTR3A and HTR3E with diarrhea predominant irritable bowel syndrome. Int J Clin Exp Med. 2015;8:4581-4585. [PubMed] |
| 17. | Kohen R, Jarrett ME, Cain KC, Jun SE, Navaja GP, Symonds S, Heitkemper MM. The serotonin transporter polymorphism rs25531 is associated with irritable bowel syndrome. Dig Dis Sci. 2009;54:2663-2670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Li Y, Hao Z, Li X, Bo P, Gong W. Association of the Serotonin Receptor 3E Gene as a Functional Variant in the MicroRNA-510 Target Site with Diarrhea Predominant Irritable Bowel Syndrome in Chinese Women. J Neurogastroenterol Motil. 2016;22:272-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: a functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Zhang Y, Huang Y, Bo P. Association between diarrhea-predominant irritable bowel syndrome and HTR3A, HTR3E gene polymorphism in Yangzhou, Jiangsu province, China. Zhonghua Liuxingbingxue Zazhi. 2013;34:721-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Bleser S. Alosetron for severe diarrhea-predominant irritable bowel syndrome: improving patient outcomes. Curr Med Res Opin. 2011;27:503-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 22. | Camilleri M, Atanasova E, Carlson PJ, Ahmad U, Kim HJ, Viramontes BE, McKinzie S, Urrutia R. Serotonin-transporter polymorphism pharmacogenetics in diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2002;123:425-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 200] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 23. | Bruder GE, Keilp JG, Xu H, Shikhman M, Schori E, Gorman JM, Gilliam TC. Catechol-O-methyltransferase (COMT) genotypes and working memory: associations with differing cognitive operations. Biol Psychiatry. 2005;58:901-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 200] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Karling P, Danielsson Å, Wikgren M, Söderström I, Del-Favero J, Adolfsson R, Norrback KF. The relationship between the val158met catechol-O-methyltransferase (COMT) polymorphism and irritable bowel syndrome. PLoS One. 2011;6:e18035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 25. | Han CJ, Kohen R, Jun S, Jarrett ME, Cain KC, Burr R, Heitkemper MM. COMT Val158Met Polymorphism and Symptom Improvement Following a Cognitively Focused Intervention for Irritable Bowel Syndrome. Nurs Res. 2017;66:75-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Ek WE, Reznichenko A, Ripke S, Niesler B, Zucchelli M, Rivera NV, Schmidt PT, Pedersen NL, Magnusson P, Talley NJ. Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut. 2015;64:1774-1782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 27. | Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut. 2013;62:985-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 28. | Weinstein LS, Chen M, Xie T, Liu J. Genetic diseases associated with heterotrimeric G proteins. Trends Pharmacol Sci. 2006;27:260-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Holtmann G, Gschossmann J, Neufang-Hüber J, Gerken G, Talley NJ. Differences in gastric mechanosensory function after repeated ramp distensions in non-consulters with dyspepsia and healthy controls. Gut. 2000;47:332-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Holtmann G, Talley NJ. Hypothesis driven research and molecular mechanisms in functional dyspepsia: the beginning of a beautiful friendship in research and practice? Am J Gastroenterol. 2006;101:593-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 31. | Wang Y, Wu Z, Qiao H, Zhang Y. A genetic association study of single nucleotide polymorphisms in GNβ3 and COMT in elderly patients with irritable bowel syndrome. Med Sci Monit. 2014;20:1246-1254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Camilleri M, Busciglio I, Carlson P, McKinzie S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Pharmacogenetics of low dose clonidine in irritable bowel syndrome. Neurogastroenterol Motil. 2009;21:399-410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Lee HJ, Lee SY, Choi JE, Kim JH, Sung IK, Park HS, Jin CJ. G protein beta3 subunit, interleukin-10, and tumor necrosis factor-alpha gene polymorphisms in Koreans with irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:758-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Markoutsaki T, Karantanos T, Gazouli M, Anagnou NP, Ladas SD, Karamanolis DG. Serotonin transporter and G protein beta 3 subunit gene polymorphisms in Greeks with irritable bowel syndrome. Dig Dis Sci. 2011;56:3276-3280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | van der Schaar PJ, van Hoboken E, Ludidi S, Masclee AA. Effect of cholecystokinin on rectal motor and sensory function in patients with irritable bowel syndrome and healthy controls. Colorectal Dis. 2013;15:e29-e34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | El-Salhy M, Gundersen D, Hatlebakk JG, Gilja OH, Hausken T. Abnormal rectal endocrine cells in patients with irritable bowel syndrome. Regul Pept. 2014;188:60-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 37. | Park SY, Rew JS, Lee SM, Ki HS, Lee KR, Cheo JH, Kim HI, Noh DY, Joo YE, Kim HS. Association of CCK(1) Receptor Gene Polymorphisms and Irritable Bowel Syndrome in Korean. J Neurogastroenterol Motil. 2010;16:71-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 38. | Cheung CK, Wu JC. Genetic polymorphism in pathogenesis of irritable bowel syndrome. World J Gastroenterol. 2014;20:17693-17698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
| 39. | Wouters MM, Lambrechts D, Knapp M, Cleynen I, Whorwell P, Agréus L, Dlugosz A, Schmidt PT, Halfvarson J, Simrén M. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut. 2014;63:1103-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 40. | Pojoga C, Stănculete MF. Biopsychosocial Approach of Gastrointestinal Disorders. Clujul Med. 2014;87:95-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 41. | Sperber AD, Dumitrascu D, Fukudo S, Gerson C, Ghoshal UC, Gwee KA, Hungin APS, Kang JY, Minhu C, Schmulson M. The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: a Rome Foundation working team literature review. Gut. 2017;66:1075-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 355] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 42. | Gazouli M, Wouters MM, Kapur-Pojskić L, Bengtson MB, Friedman E, Nikčević G, Demetriou CA, Mulak A, Santos J, Niesler B. Lessons learned--resolving the enigma of genetic factors in IBS. Nat Rev Gastroenterol Hepatol. 2016;13:77-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 43. | Boeckxstaens GE, Drug V, Dumitrascu D, Farmer AD, Hammer J, Hausken T, Niesler B, Pohl D, Pojskic L, Polster A. Phenotyping of subjects for large scale studies on patients with IBS. Neurogastroenterol Motil. 2016;28:1134-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 44. | Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 45. | Kilpatrick LA, Labus JS, Coveleskie K, Hammer C, Rappold G, Tillisch K, Bueller JA, Suyenobu B, Jarcho JM, McRoberts JA. The HTR3A polymorphism c. -42C> T is associated with amygdala responsiveness in patients with irritable bowel syndrome. Gastroenterology. 2011;140:1943-1951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |