Published online Oct 26, 2015. doi: 10.13105/wjma.v3.i5.215
Peer-review started: January 10, 2015
First decision: June 3, 2015
Revised: June 26, 2015
Accepted: July 24, 2015
Article in press: July 27, 2015
Published online: October 26, 2015
Processing time: 294 Days and 21.6 Hours
AIM: To compare four methods to approximate mean and standard deviation (SD) when only medians and interquartile ranges are provided.
METHODS: We performed simulated meta-analyses on six datasets of 15, 30, 50, 100, 500, and 1000 trials, respectively. Subjects were iteratively generated from one of the following seven scenarios: five theoretical continuous distributions [Normal, Normal (0, 1), Gamma, Exponential, and Bimodal] and two real-life distributions of intensive care unit stay and hospital stay. For each simulation, we calculated the pooled estimates assembling the study-specific medians and SD approximations: Conservative SD, less conservative SD, mean SD, or interquartile range. We provided a graphical evaluation of the standardized differences. To show which imputation method produced the best estimate, we ranked those differences and calculated the rate at which each estimate appeared as the best, second-best, third-best, or fourth-best.
RESULTS: Our results demonstrated that the best pooled estimate for the overall mean and SD was provided by the median and interquartile range (mean standardized estimates: 4.5 ± 2.2, P = 0.14) or by the median and the SD conservative estimate (mean standardized estimates: 4.5 ± 3.5, P = 0.13). The less conservative approximation of SD appeared to be the worst method, exhibiting a significant difference from the reference method at the 90% confidence level. The method that ranked first most frequently is the interquartile range method (23/42 = 55%), particularly when data were generated according to the Standard Normal, Gamma, and Exponential distributions. The second-best is the conservative SD method (15/42 = 36%), particularly for data from a bimodal distribution and for the intensive care unit stay variable.
CONCLUSION: Meta-analytic estimates are not significantly affected by approximating the missing values of mean and SD with the correspondent values for median and interquartile range.
Core tip: Meta-analyses of continuous endpoints are generally supposed to deal with normally distributed data and the pooled estimate of the treatment effect relies on means and standard deviations. However, if the outcome distribution is skewed, some authors correctly report the median together with the corresponding quartiles. In the present work, we compared methods for the approximation of means and standard deviations when only medians with quartiles are provided. Our results demonstrate that meta-analytic estimates are not significantly affected by approximating the missing values of mean and standard deviation with the correspondent values for median and interquartile range.
- Citation: Greco T, Biondi-Zoccai G, Gemma M, Guérin C, Zangrillo A, Landoni G. How to impute study-specific standard deviations in meta-analyses of skewed continuous endpoints? World J Meta-Anal 2015; 3(5): 215-224
- URL: https://www.wjgnet.com/2308-3840/full/v3/i5/215.htm
- DOI: https://dx.doi.org/10.13105/wjma.v3.i5.215
Meta-analysis (MA) is a powerful statistical method that merges the results of different studies considering the same outcome variables. The included studies are mainly randomized controlled trials with experimental and control arms. MA aims at assessing the treatment effect size under scrutiny, at identifying sources of heterogeneity among the included studies, at unrevealing patterns behind the available data, and sometimes at identifying new subgroup associations. Some authors believe that MA represents the highest level of evidence to provide recommendations on clinical issues.
Just as many other innovative statistical techniques, MA is still a matter of intense debate, since many of its assumptions are critical and even small violations of them can lead to misleading conclusions[1]. The appeal of MA resides also in the quick and cost-effective way it yields useful pieces of information for clinical decision making. Hence, MAs are published and quoted with an impressive increasing frequency and it seems evident that, despite their limitations, they will continue to play a crucial role in medical decision-making in the foreseeable future.
Meta-analyses of continuous outcomes exploit data with a Gaussian distribution, so that the pooled estimate computation requires the study-specific mean, standard deviation (SD), and sample size of the variable at stake. The easiest way to compare the outcomes of two treatment groups is to evaluate the difference between their means[2]. If measurements are expressed in the same unit, the mean difference between the treatment and control groups can be used. Results from trials in which the same outcome is measured in different units can be compared by using SD units rather than absolute differences[2,3]. However, if data are reported in a limited or incomplete way, it can be difficult or impossible to obtain sufficient information to perform a correct summary of the results. Missing SD and non-compliance in reporting collected data are common limitations in MAs of continuous outcomes.
If the outcome has a skewed distribution, authors often report in the original paper the median together with the 1st and 3rd quartiles, rather than the mean with its SD. MA authors arbitrarily combine these study-specific estimates to approximate the missing SD. In addition, some authors combine together means (SD) and medians (1st and 3rd quartiles) from different studies.
In this setting, the most immediate question is whether it is legitimate to approximate study-specific means and SDs from study-specific medians and quartiles and how to do it in the most appropriate way.
In the present work we simulated MA of continuous outcomes generated from seven different distributions and we compared four methods to approximate SDs when only study-specific medians and quartiles are available. For each simulation we calculated a pooled estimate by assembling the individual medians and, in turn, all four SD approximations. Finally, we compared these results with those obtained by pooling the individual means and SDs.
To our knowledge, this is the first study on how to impute the study-specific mean and SD in meta-analyses of skewed outcomes. After a failed tentative of comparing results coming from published studies, the present paper compares the available methods making use of both simulations and real-life set-ups to identify the best and the worst SD approximation.
We generated six datasets of 15, 30, 50, 100, 500, and 1000 trials, respectively. Each trial is based on an equal number of treated and control subjects which is fixed at the number of trials included in the MA under examination. The distributions of the continuous endpoints for the treatment and control groups are generated according to Table 1. The first five scenarios provided the basis for our analysis on simulated data. The last two scenarios of Table 1 represent our real-life data and are randomly extracted from an Italian observational study with more than 7000 patients with cardiovascular disease.
| Scenario | Endpoint distribution | |
| Treatment group | Control group | |
| Normal | Mean = 5 and SD = 2 | Mean = 7 and SD = 2 |
| Standard normal | Mean = 0 and SD = 1 | Mean = 0 and SD = 1 |
| Gamma | Alpha = 2 and beta = 5 | Alpha = 2 and beta = 7 |
| Exponential | Mean = 5 and lambda = 0.2 | Mean = 7 and lambda = 0.14 |
| Bimodal | 50% Normal distribution with mean = 5 and SD = 2 and 50% standard normal distribution | 50% Normal distribution with mean = 7 and SD = 2 and 50% standard normal distribution |
| ICU stay | Real-life data | Real-life data |
| Hospital stay | Real-life data | Real-life data |
In summary, we generated 1695 (15 + 30 + 50 + 100 + 500 + 1000) trials for each of the seven distribution scenarios for a total of 11865 trials. For each trial we calculated the principal measures of position, mean and median, and variability, SD and interquartile range (IQR).
All simulations and analyses were performed using SAS (release 9.2, 2002-2008 by SAS Institute Inc., Cary, NC, United States)[4]. An example of SAS code is reported in Table 2.
| * q is the assigned library; |
| **************************************************; |
| * SIMULATIONS; |
| **************************************************; |
| %let s = gamma; |
| %let ndset = 15; |
| * Simulation of n = 15 dataset using the Gamma distributions; |
| %macro simul; |
| %do q = 1 %to &ndset; |
| %let seed = %sysevalf(1234567 + &q); |
| %let num_i = %sysevalf(&ndset); |
| %let v = %sysevalf(0 + &q); |
| data s&q; |
| k = &q; |
| %do i = 1%to &num_i; |
| var1 = 5*rangam(&seed,2); |
| var2 = 7*rangam(&seed,2); |
| output; |
| %end; |
| run; |
| %end; |
| * Dataset combining; |
| data simul_&s; |
| set |
| %do w = 1%to &ndset; |
| s&w |
| %end; |
| ; |
| run; |
| %mend; |
| %simul; |
| * Descriptive statistics for each dataset; |
| ods trace on; |
| ods output summary = summary_&s; |
| proc means data = simul_&s mean std median q1 q3; |
| class k; |
| var var1 var2; |
| run; |
| ods trace off; |
| data summary_&s; |
| set summary_&s; |
| l1 = (var1_Median-var1_Q1)/0.6745; |
| l2 = (var2_Median-var2_Q1)/0.6745; |
| u1 = (var1_Q3-var1_Median)/0.6745; |
| u2 = (var2_Q3-var2_Median)/0.6745; |
| if l1 > u1 then MeSD_v1_cons=l1; else MeSD_v1_cons=u1; |
| if l2 > u2 then MeSD_v2_cons=l2; else MeSD_v2_cons=u2; |
| if l1 > u1 then MeSD_v1_prec=u1; else MeSD_v1_prec=l1; |
| if l2 > u2 then MeSD_v2_prec=u2; else MeSD_v2_prec=l2; |
| MeSD_v1_mean=(var1_Q3-var1_Q1)/1.349; |
| MeSD_v2_mean=(var2_Q3-var2_Q1)/1.349; |
| * Median difference; |
| MeD = var1_Median-var2_Median; |
| *1 conservative estimate of standard deviation; |
| a1sd = ((MeSD_v1_cons)**2)/NObs; |
| b1sd = ((MeSD_v2_cons)**2)/NObs; |
| MeSD_cons=sqrt(a1sd + b1sd); |
| *2 less conservative estimate of standard deviation; |
| a2sd = ((MeSD_v1_prec)**2)/NObs; |
| b2sd = ((MeSD_v2_prec)**2)/NObs; |
| MeSD_prec = sqrt(a2sd + b2sd); |
| *3 mean estimate of standard deviation; |
| a3sd = ((MeSD_v1_mean)**2)/NObs; |
| b3sd = ((MeSD_v2_mean)**2)/NObs; |
| MeSD_mean = sqrt(a3sd + b3sd); |
| *4 Interquartile range; |
| a4sd = ((var1_Q3-var1_Q1)**2)/NObs; |
| b4sd = ((var2_Q3-var2_Q1)**2)/NObs; |
| MeSD_iqr = sqrt(a4sd + b4sd); |
| * Mean difference and pooled standard deviation; |
| MD = var1_Mean-var2_Mean; |
| asd = ((var1_StdDev)**2)/NObs; |
| bsd = ((var2_StdDev)**2)/NObs; |
| SD = sqrt(asd + bsd); |
| drop l1 l2 u1 u2 asd bsd a1sd b1sd a2sd b2sd a3sd b3sd a4sd b4sd; |
| run; |
| *************************; |
| * Meta-analyses; |
| data sum_&s; |
| set summary_&s; |
| keep k NObs MeD MeSD_cons MeSD_prec MeSD_mean MeSD_iqr MD SD qq; |
| run; |
| *1 Median and conservative estimate of standard deviation; |
| data meta_&s.1; |
| set sum_&s; |
| model = "Conservative SD"; |
| MDz = MeD; |
| SDz = MeSD_cons; |
| w = 1/(SDz**2); |
| MDw = MDz*w; |
| keep model k NObs MDz SDz w MDw; |
| run; |
| *2 Median and less conservative estimate of standard deviation; |
| data meta_&s.2; |
| set sum_&s; |
| model = "Less Conservative SD"; |
| MDz = MeD; |
| SDz = MeSD_prec; |
| w = 1/(SDz**2); |
| MDw = MDz*w; |
| keep model k NObs MDz SDz w MDw; |
| run; |
| *3 Median and mean estimate of standard deviation; |
| data meta_&s.3; |
| set sum_&s; |
| model = "Mean SD"; |
| MDz = MeD; |
| SDz = MeSD_mean; |
| w = 1/(SDz**2); |
| MDw=MDz*w; |
| keep model k NObs MDz SDz w MDw; |
| run; |
| *4 Median and interquartile range; |
| data meta_&s.4; |
| set sum_&s; |
| model = "IQR"; |
| MDz = MeD; |
| SDz = MeSD_iqr; |
| w = 1/(SDz**2); |
| MDw = MDz*w; |
| keep model k NObs MDz SDz w MDw; |
| run; |
| *Mean and standard deviation (reference); |
| data meta_&s.5; |
| set sum_&s; |
| model = "Reference"; |
| MDz = MD; |
| SDz = SD; |
| w = 1/(SDz**2); |
| MDw = MDz*w; |
| keep model k NObs MDz SDz w MDw; |
| run; |
| proc format; |
| value model |
| 1 = "conservative SD" |
| 2 = "Less Conservative SD " |
| 3 = "Mean SD " |
| 4 = "IQR" |
| 5 = "Reference" |
| ; |
| run; |
| *** Fixed effect model meta-analysis - Inverse of Variance method; |
| %macro meta_iv; |
| %do i = 1%to 5; |
| ods output Summary = somme&i; |
| proc means data = meta_&s&i sum; |
| var MDw w; |
| run; |
| data somme&i; |
| set somme&i; |
| model = &i; |
| format model model.; |
| theta = MDw_Sum/w_Sum; |
| se_theta = 1/(sqrt(w_sum)); |
| lower = theta - (se_theta*1.96); |
| upper = theta + (se_theta*1.96); |
| mtheta = sqrt(theta**2); |
| CV = se_theta/mtheta; |
| keep model theta se_theta lower upper cv; |
| run; |
| %end; |
| data aaMeta_&s; |
| set |
| %do w = 1% to 5; |
| somme&w |
| %end; |
| ; |
| run; |
| title "distr = &s - k = &ndset"; |
| proc print; run; |
| %mend; |
| %meta_iv; |
For each of the six datasets we carried out a series of MAs differing with respect to the method of imputation of the study-specific SD[2], as described in Table 3.
| Methodnumber | Method name | Mean imputation | Standard Deviation imputation1 |
| 0 | Reference | Mean | SD |
| 1 | Conservative SD | Median | max[(3rd quartile - median)/0.6745; (median - 1st quartile)/0.6745] |
| 2 | Less Conservative SD | Median | min[(3rd quartile - median)/0.6745; (median - 1st quartile)/0.6745] |
| 3 | Mean SD | Median | (3rd quartile - 1st quartile)/(2 × 0.6745) |
| 4 | IQR | Median | (3rd quartile - 1st quartile) |
For each distribution scenario, 30 MAs were therefore performed (6 datasets × 5 methods).
Each MA was carried out using a fixed effect model by the Inverse-Variance method[3].
The global estimate is the mean difference between treatment and control groups, obtained by pooling individual means and SDs.
For each distribution scenario we computed four standardized estimates, θstandijk, calculated as:
θstandijk = (θijk - θreferenceijk)/ se(θijk)
for
i = 1, 2, 3, 4; j = 15, 30,..., 1000 and k = 1, 2,…, 7.
where θijk is the pooled mean difference resulting from the performed MA, se(θijk) is the corresponding standard error and θreferenceijk is the global estimate obtained by pooling individual means and SDs for each dataset and distribution scenario.
We obtained a total of 168 standardized estimates (7 distributions × 6 datasets × 4 methods).
After blocking for dataset and distribution, we evaluated if the standardized estimates, calculated by each of the four methods, were different from zero in the framework of a repeated measures model using the mixed procedure implemented in the SAS software. Statistical significance was set at the two-tailed 0.05 level for hypothesis testing. Adjustment for multiple comparisons was performed with Tukey-Kramer, Bonferroni and Scheffé corrections[5,6].
To identify which imputation method produced the pooled estimate θijk with the minimum difference from the reference one, θreferenceijk, we ranked each standardized estimate θstandijk for each dataset and for each distribution scenario. According to this ranking, we calculated the rate at which each estimate appeared as the best, second-best, third-best, or fourth-best[7] and the area under the cumulative ranking curve[8].
We screened all the best published studies reported in the medical literature on critically ill patients included in the 1st web based International Consensus Conference[9] to assess which information authors of original papers provided in the case of intensive care unit (ICU) and hospital stay outcome. We contacted the corresponding authors by e-mail when one or more of the following information was missing: Mean, SD, median, 1st-3rd quartiles. No manuscript provided all of the information requested and only 3 authors replied to our e-mail and provided the extra information needed.
The statistical methods of this study were reviewed by Rosalba Lembo from San Raffaele Scientific Institute, Via Olgettina 60, 20132 Milan, Italy.
RESULTS
Table 4 reports the means and the corresponding unadjusted and adjusted P values to assess the presence of a significant difference from zero. Conservative SD and IQR methods showed the lowest mean difference (with mean standardized estimates equal to 4.5 ± 3.5 for conservative SD and 4.5 ± 2.2 for IQR methods, respectively). The less conservative SD method appeared to be the worst, exhibiting the highest difference from the reference.
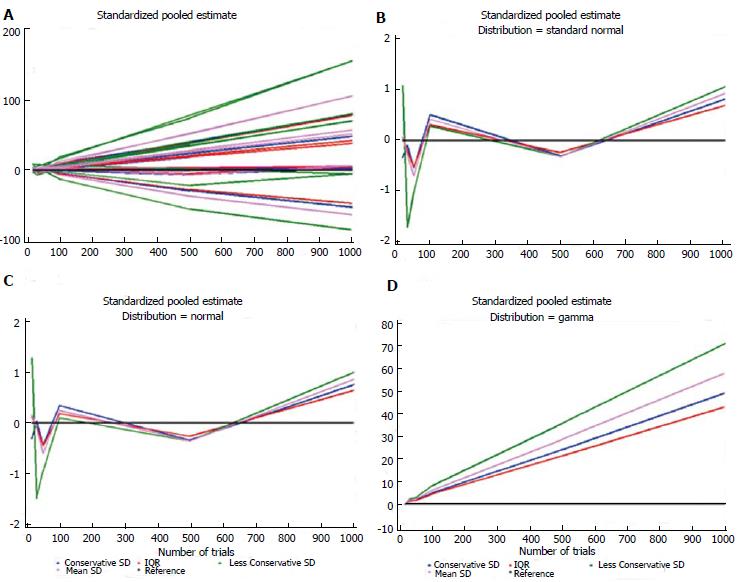
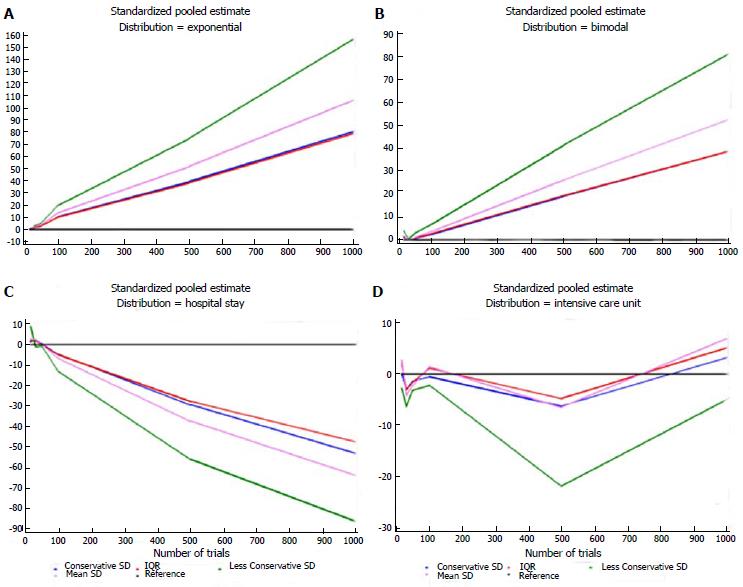
Table 5 shows the estimates for each distribution, dataset, and imputation method and some descriptive statistics. For each distribution, the number of times each standardized estimate ranked first is indicated. The method that ranked first most frequently was IQR (23/42 = 55%), particularly when the data were generated according to the Standard Normal, Gamma, and Exponential distributions. The second best is the Conservative SD method (15/42 = 36%), which was particularly suitable for data with a bimodal distribution and for the ICU stay variable. The quartiles at the bottom of Table 5 were similar for these two methods: 1.62 (0.27-4.94) and 1.40 (0.34-4.86), respectively. Figures 1 and 2 show plots of the pooled estimates for all distribution scenarios. The difference between θstandijk and reference values increased together with the increased number of trials in each MA.
| Distributionscenario | Dataset | Conservative SD | Less Conservative SD | Mean SD | IQR |
| Normal | 15 | -0.310 | 1.274 | 0.149 | 0.110 |
| Normal | 30 | 0.029 | -1.483 | -0.109 | -0.081 |
| Normal | 50 | -0.434 | -0.946 | -0.599 | -0.444 |
| Normal | 100 | 0.340 | 0.095 | 0.243 | 0.180 |
| Normal | 500 | -0.336 | -0.357 | -0.353 | -0.261 |
| Normal | 1000 | 0.754 | 0.989 | 0.860 | 0.638 |
| No. of times of beginning first in the ranking1 | 2 | 1 | 0 | 3 | |
| Standard normal | 15 | -0.335 | 1.072 | 0.062 | 0.046 |
| Standard normal | 30 | -0.101 | -1.710 | -0.290 | -0.215 |
| Standard normal | 50 | -0.535 | -1.013 | -0.690 | -0.511 |
| Standard normal | 100 | 0.502 | 0.281 | 0.416 | 0.308 |
| Standard normal | 500 | -0.306 | -0.314 | -0.317 | -0.235 |
| Standard Normal | 1000 | 0.814 | 1.054 | 0.923 | 0.684 |
| No. of times of beginning first in the ranking1 | 1 | 1 | 0 | 4 | |
| Gamma | 15 | -0.283 | 0.054 | -0.119 | -0.088 |
| Gamma | 30 | 1.441 | 2.229 | 1.846 | 1.368 |
| Gamma | 50 | 1.795 | 2.929 | 2.218 | 1.644 |
| Gamma | 100 | 4.915 | 8.193 | 6.070 | 4.500 |
| Gamma | 500 | 24.081 | 35.799 | 28.753 | 21.314 |
| Gamma | 1000 | 49.089 | 71.072 | 58.012 | 43.002 |
| No. of times of beginning first in the ranking1 | 0 | 1 | 0 | 5 | |
| Exponential | 15 | -0.150 | -0.163 | -0.157 | -0.116 |
| Exponential | 30 | 1.880 | 2.958 | 2.301 | 1.706 |
| Exponential | 50 | 2.948 | 4.975 | 3.707 | 2.748 |
| Exponential | 100 | 10.213 | 19.490 | 13.493 | 10.002 |
| Exponential | 500 | 39.546 | 74.955 | 51.913 | 38.481 |
| Exponential | 1000 | 80.605 | 157.083 | 106.593 | 79.016 |
| No. of times of beginning first in the ranking1 | 0 | 0 | 0 | 6 | |
| Bimodal | 15 | 1.142 | 4.114 | 1.751 | 1.298 |
| Bimodal | 30 | 0.079 | 0.356 | 0.096 | 0.071 |
| Bimodal | 50 | 0.545 | 3.051 | 1.110 | 0.823 |
| Bimodal | 100 | 2.405 | 6.849 | 3.650 | 2.706 |
| Bimodal | 500 | 19.156 | 41.495 | 26.212 | 19.431 |
| Bimodal | 1000 | 38.825 | 81.301 | 52.527 | 38.938 |
| No. of times of beginning first in the ranking1 | 5 | 0 | 0 | 1 | |
| ICU stay | 15 | 0.076 | -2.816 | 2.667 | 1.977 |
| ICU stay | 30 | -3.011 | -6.341 | -4.201 | -3.114 |
| ICU stay | 50 | -1.361 | -3.162 | -2.163 | -1.603 |
| ICU stay | 100 | -0.58 | -2.205 | 1.393 | 1.032 |
| ICU stay | 500 | -6.218 | -21.788 | -6.462 | -4.790 |
| ICU stay | 1000 | 3.162 | -5.020 | 6.801 | 5.042 |
| No. of times of beginning first in the ranking1 | 5 | 0 | 0 | 1 | |
| Hospital stay | 15 | 1.437 | 8.948 | 2.777 | 2.058 |
| Hospital stay | 30 | 2.603 | -1.088 | 2.595 | 1.924 |
| Hospital stay | 50 | 0.297 | -0.839 | -0.055 | -0.041 |
| Hospital stay | 100 | -4.674 | -13.063 | -6.734 | -4.992 |
| Hospital stay | 500 | -29.239 | -55.703 | -37.170 | -27.554 |
| Hospital stay | 1000 | -52.720 | -85.673 | -63.453 | -47.038 |
| No. of times of beginning first in the ranking1 | 2 | 1 | 0 | 3 | |
| 1st quartile | 0.337 | 1.023 | 0.368 | 0.273 | |
| Median | 1.399 | 2.944 | 2.190 | 1.624 | |
| 3rd quartile | 4.855 | 12.034 | 6.666 | 4.942 | |
| Total number of times of beginning first in the ranking1 | 15 | 4 | 0 | 23 |
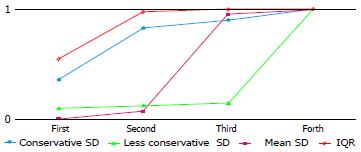
Table 6 reports the rates of occurrence as best, second-best, third-best and fourth-best, for the four imputation methods. The conservative SD and IQR methods most often appeared as best or second-best (cumulative frequencies: 35/42 and 41/42 respectively), and the less conservative SD and mean SD methods as third- or fourth-best. The less conservative SD method was identified as the worst method as it had by far the highest number of fourth positions. Even when it ranked first, the θstandijk was very similar to the IQR method, which is consistently suitable for any distribution scenario.
| No. of firstranking | No. of second ranking | No. of third ranking | No. of fourth ranking | |
| Conservative SD | 15 (35.7) | 20 (47.6) | 3 (7.1) | 4 (9.5) |
| Less Conservative SD | 4 (9.5) | 1 (2.4) | 1 (2.4) | 36 (85.7) |
| Mean SD | 0 | 3 (7.1) | 37 (88.1) | 2 (4.8) |
| IQR | 23 (54.8) | 18 (42.9) | 1 (2.4) | 0 |
Figure 3 shows the areas under the cumulative ranking curve for each imputation method. IQR method yields the largest area.
The aim of our work was to clarify how to best impute study-specific mean and SD when only the median and 1st and 3rd quartiles are provided.
It is a notoriously good practice to report information on medians and quartiles for skewed distributions but standard meta-analytic approaches require study-specific means and SDs, so that careful evaluation of the best trade-off between these two approaches is needed.
This issue is of prominent interest when dealing with MA, but few scientific works addressed this topic. Hozo et al[10] described two formulas for estimating the mean from the median, range, and sample size values. Pigott[11] discussed and examined how to deal with missing data (studies, effect size, and methodological information) during MA. Furukawa et al[12,13] reported on the imputation of the missing response rate from the mean ± SD[12] and suggested that borrowing the missing SD from other studies included in the MA may be a valid solution[13]. Thiessen Philbrook et al[14] compared results from MAs that either were restricted to available data or imputed the missing variance with one of four methods (P values, nonparametric summaries, multiple imputation, or correlation coefficients). Robertson et al[15] highlighted and evaluated different ways to include in MAs studies in which the treatment effect was not provided. Weibe et al[16] conducted a systematic review of methods for handling missing variances in MAs of continuous outcomes and classified the relevant approaches into eight groups: algebraic recalculation, approximate algebraic recalculation, study-level imputation, study-level imputation from nonparametric summaries, study-level imputation of correlation (for change-from-baseline or crossover SD and to calculate the design effect for cluster studies), MA-level imputation of overall effect, MA-level tests, and no-imputation methods. Finally, Stevens[17] gave an overview of the Bayesian approach to deal with missing data in MA. However, authors who carry out MAs rarely adopt similar methods in their current clinical practice.
In the present study, we showed that MA pooled estimates are not significantly affected by approximating the missing study-specific mean and SD with the corresponding median and IQR, in both simulated and real-life set-ups. In comparison with the other methods, we found that the Median-IQR method has extra advantages, since it is the simplest one and it makes no assumption on the underlying distribution of the data. Furthermore, we showed how the use of a less conservative approximation of SD can bias the meta-analytic pooled estimate when authors work with skewed data. Nevertheless, it is well known that median and mean values are very different as the data distribution is skewed[2].
Our study has several limitations. First, we did not perform a sensitivity analysis with the random effects model. However, since data were generated from the same distribution for each trial, we decided to apply fixed effect models. Second, we did not analyze mixed set-ups in which study-specific means and SDs are available for some studies and others are imputed. Third, we worked only on trials where the number of treated/control subjects was equal to the number of studies included in the MA. It follows that we did not consider set-ups with many studies and few subjects or vice versa. Although we present several distribution scenarios, the choice of their parameters was arbitrary. However, the performance is not expected to change with different parameters.
It is not surprising that many papers report median and IQR, rather than mean and SD. Actually this is considered good practice when dealing with non-normally distributed data. As an example, since the distributions of ICU or hospital stay are skewed, it might be worth using median and IQR when setting up any MA with these end-points.
Our work supports the procedure of using study-specific medians and quartiles to impute means and standard deviations. This avoids the dangerous practice of not including in the MA studies with missing information. Nevertheless, we recognize that, in order to improve the quality of future MAs, authors of research papers should report as much information as possible at least for concerning their primary outcomes. We suggest that authors who use median and interquartile range in MAs with continuous endpoints, perform a sensitivity analysis in which trials not providing study-specific means and SDs are excluded.
Meta-analyses (MAs) of continuous outcomes exploit data with a Gaussian distribution, so that the pooled estimate computation requires the study-specific means, standard deviations (SDs), and sample sizes. However, should data be reported in a limited or incomplete way, it can be difficult or impossible to obtain sufficient information to perform a correct summary of the results. Missing standard deviations and non-compliance in reporting collected data are common limitations in MAs of continuous outcomes. At the simpler level, an interest emerged as to the opportunity and the best way to approximate on study-specific means and SDs from study-specific medians and quartiles.
The aim of the work was to clarify how to best impute study-specific mean and SD when only the median and 1st and 3rd quartiles are provided. The authors compared the available methods in simulated and real-life set-ups to identify the best and the worst one. In the present study, authors showed that MA pooled estimates are not significantly affected by approximating the missing study-specific mean and SD with the corresponding median and interquartile range. Among the four methods proposed, the Median-IQR method has the extra advantages since it is the simplest one and that makes no assumption on the underlying distribution of the data.
For the first time, to our knowledge, this manuscript provides a list of four available approximation methods in MAs of skewed outcomes. The authors became aware of these methods in the clinical practice and we strongly believe in the good practice to report information on medians and quartiles when a distribution is skewed. However, standard meta-analytic approaches require study specific means and SDs and it was natural to us to find out a compromise between these two needs. This issue is important when carrying out a MA, but few scientific works addressed this topic. Authors described formulas and estimating procedures to work with missing data in an MA, in either frequentist or Bayesian approach. However, authors who carry out MAs rarely adopt similar methods in their current clinical practice.
The work gives support to the procedure of using study-specific medians and quartiles to impute means and SDs in an MA of skewed outcome. This avoids the dangerous practice of not including in the MA studies with missing information.
Approximation: An estimate of the value of a quantity to a desired degree of accuracy; Conservative estimate: Estimate that avoids excess in approximating the quantity or worth of the list of (potentially infinite) values identified; Distribution: List of the values in a population, or sample, with the corresponding frequency or probability of occurrence; Estimate: A value (a point estimate) or range of values (an interval estimate) to a parameter of a population on the basis of sampling statistics; Interquartile range: Measure of dispersion around the median given by the difference between the 3rd quartile and 1st quartile of the distribution; these quartiles can be clearly seen on a box-plot of the data; Study-specific: Related to a specific study included in a more comprehensive MA (i.e., group of study).
The study addresses a very interesting topic and deals with a major concern for investigators who have to perform a meta-analysis from published studies.
P- Reviewer: Naugler C, Omboni S, Trkulja V S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | Greco T, Zangrillo A, Biondi-Zoccai G, Landoni G. Meta-analysis: pitfalls and hints. Heart Lung Vessel. 2013;5:219-225. [PubMed] |
| 2. | Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions. Version 5.1.0. The Cochrane collaboration. 2011;[Accessed 2015 Jun] Available from: http://www.cochranehandbook.org/. |
| 3. | Rothstein HR, Sutton AJ, Borenstein M. Publication Bias in Meta-Analysis: Prevention, Assessment and Adjustments. Chapter 8. The Trim and Fill Method. New York: John Wiley & Sons Ltd 2006; . [DOI] [Full Text] |
| 4. | Moser EB. Repeated measures modeling with PROC MIXED. In: Proceedings of the 29th SAS Users Group International Conference. Montreal, Canada 2004. [Accessed 2015; Jun] Available from: http://www2.sas.com/proceedings/sugi29/188-29.pdf. |
| 5. | Edwards D, Berry JJ. The efficiency of simulation-based multiple comparisons. Biometrics. 1987;43:913-928. [PubMed] |
| 6. | Rafter JA, Abell ML, Braselton JP. Multiple Comparison Methods for Means. SIAM Review. 2002;44:259-278. |
| 7. | Dias S, Welton NJ, Sutton AJ, Ades AE. NICE DSU Technical Support Document2: A generalised linear modelling framework for pairwise and network meta-analysis of randomised controlled trials 2011 [Accessed 2015 Jun]. Available from: http://www.nicedsu.org.uk. |
| 8. | Flach P, Matsubara ET. On classification, ranking, and probability estimation. Dagstuhl Seminar Proceedings 07161. Probabilistic, Logical and Relational Learning - A Further Synthesis. In: Proceedings of the 29th SAS Users Group International Conference. Montreal, Canada 2004. [Accessed 2008; [Accessed 2015 Jun] Available from: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.183.6043&rep=rep1&type=pdf. |
| 9. | Landoni G, Augoustides JG, Guarracino F, Santini F, Ponschab M, Pasero D, Rodseth RN, Biondi-Zoccai G, Silvay G, Salvi L. Mortality reduction in cardiac anesthesia and intensive care: results of the first International Consensus Conference. Acta Anaesthesiol Scand. 2011;55:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5:13. [PubMed] |
| 11. | Pigott TD. Missing predictors in models of effect size. Eval Health Prof. 2001;24:277-307. |
| 12. | Furukawa TA, Cipriani A, Barbui C, Brambilla P, Watanabe N. Imputing response rates from means and standard deviations in meta-analyses. Int Clin Psychopharmacol. 2005;20:49-52. [PubMed] |
| 13. | Furukawa TA, Barbui C, Cipriani A, Brambilla P, Watanabe N. Imputing missing standard deviations in meta-analyses can provide accurate results. J Clin Epidemiol. 2006;59:7-10. [PubMed] |
| 14. | Thiessen Philbrook H, Barrowman N, Garg AX. Imputing variance estimates do not alter the conclusions of a meta-analysis with continuous outcomes: a case study of changes in renal function after living kidney donation. J Clin Epidemiol. 2007;60:228-240. [PubMed] |
| 15. | Robertson C, Idris NR, Boyle P. Beyond classical meta-analysis: can inadequately reported studies be included? Drug Discov Today. 2004;9:924-931. [PubMed] |
| 16. | Wiebe N, Vandermeer B, Platt RW, Klassen TP, Moher D, Barrowman NJ. A systematic review identifies a lack of standardization in methods for handling missing variance data. J Clin Epidemiol. 2006;59:342-353. [PubMed] |
| 17. | Stevens JW. A note on dealing with missing standard errors in meta-analyses of continuous outcome measures in WinBUGS. Pharm Stat. 2011;10:374-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |