Copyright
©The Author(s) 2016.
World J Transplant. Mar 24, 2016; 6(1): 28-41
Published online Mar 24, 2016. doi: 10.5500/wjt.v6.i1.28
Published online Mar 24, 2016. doi: 10.5500/wjt.v6.i1.28
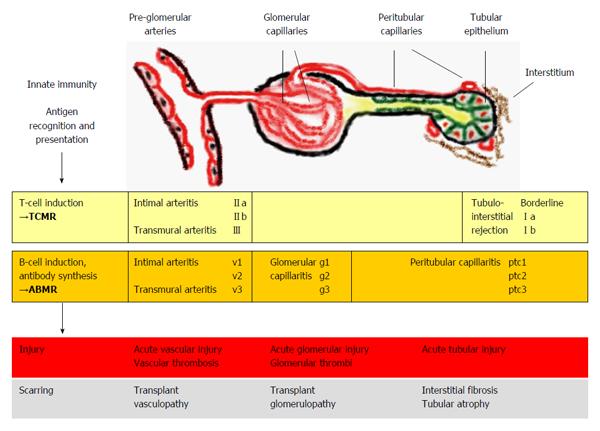
Figure 1 Kidney allograft rejection types, histological sites of injury and underlying mechanisms.
TCMR includes recognition and presentation of donor antigens by antigen-presenting cells to T cells, which become activated and undergo proliferation. Activated T-cells invade vascular, tubular and interstitial structures. Vascular rejection often presents with some degree of tubulointerstitial inflammation; however pure cases of vascular rejection (“v-only”) can be observed[8]. In ABMR, activated T cells induce B cells to undergo plasma cell proliferation resulting in the production of donor-specific antibodies. Antibody-mediated injury to pre-glomerular arteries, glomerular and peritubular capillaries is mediated by local activation of complement factors however, non-complement-fixing antibodies may also play a role in some cases[9]. Isolated findings of glomerular and peritubular capillaritis or pre-glomerular arteritis may be present or a combination of these features[7]. TCMR and ABMR can occur simultaneously (i.e., mixed rejection)[4]. The rejection processes can lead to different histological forms of injury and if not successfully treated, to scarring. The Banff classification[7] associates the elementary lesions of glomerular (g) and peritubular capillaries (ptc) and pre-glomerular vessels (v) to ABMR. TCMR includes tubulointerstitial infiltration (Borderline, I) and arteritis of pre-glomerular vessels (II-III). Banff grades (a-b, II-III, v1-3, g1-3, ptc1-3) denote different severities of the lesions. TCMR: T cell-mediated rejection; ABMR: Antibody-mediated rejection.
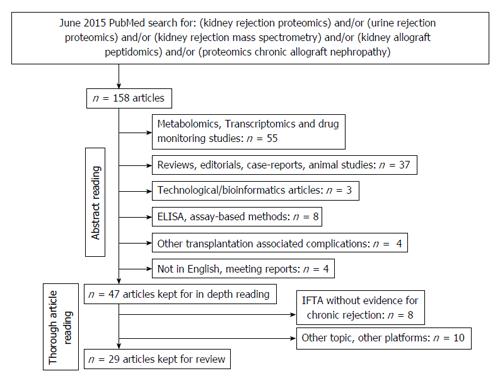
Figure 2 Search strategy for proteomic studies in the field of renal allograft rejection.
IFTA: Interstitial fibrosis and tubular atrophy.
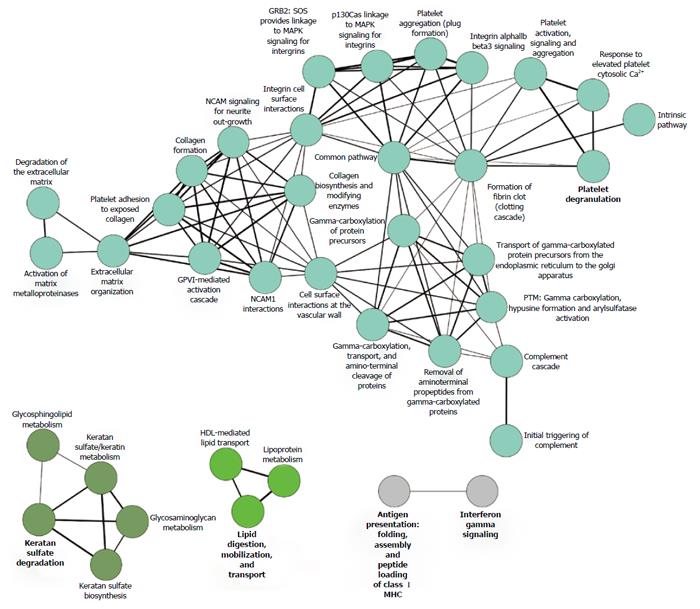
Figure 3 Reactome graph, showing the functional association of renal allograft rejection molecules.
Literature-derived proteins associated with acute and chronic rejection (n = 89, concatenated from the proteomic studies listed in Table 1) were analyzed by functional Reactome group-clustering using CytoScape’s ClueGO plug-in (CytoScape v2.8.3, ClueGO v1.5). Enriched Reactome-terms are represented as circles, and lines denote the relationship between these terms as functional groups. Line thickness and font-size are directly correlated with the statistical significance of terms and relationships (all with P < 0.05 after Bonferroni-adjustment for multiple testing correction). MAPK: Mitogen-activated protein kinase; GRB2: Growth factor receptor-bound protein 2; NCAM: Neural cell adhesion molecule.
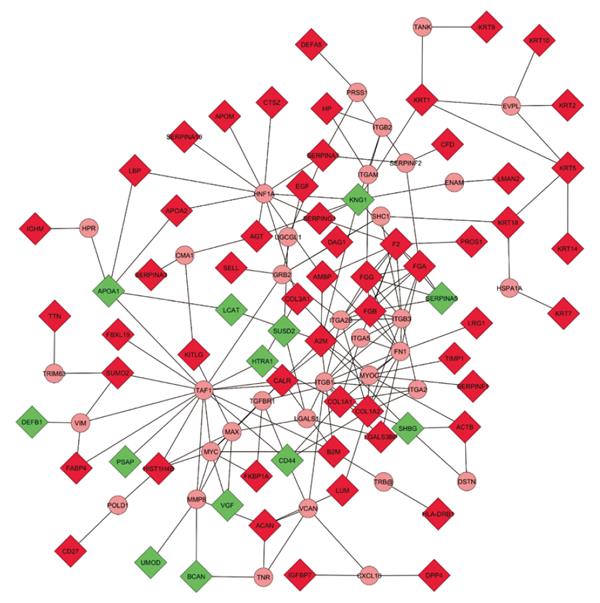
Figure 4 Expanded molecular interaction model.
Physical interaction representation of molecules involved in renal allograft rejection. The concatenated list of literature-derived proteins associated with acute and chronic rejection was subjected to interactome-mapping using CytoScape’s Michigan Molecular Interactor (MiMI) plug-in (CytoScape v2.8.2, MiMI v3.1). Known protein-protein interactions with up to two additional bridging molecules to maximize the interconnectivity were used to generate the map shown, which contains 68 of the 89 differentially expressed molecules and 35 additional bridging proteins. Input molecules are depicted as rectangles, and bridging molecules as circles. Each line between proteins represents a direct known association. Included literature-derived proteins associated with acute and chronic renal allograft rejection in the network (Rectangles; Green: Down-regulated; Red: Up-regulated; n = 68); Included additional bridging proteins for maximum interconnectivity (circles; n = 35); Excluded literature-derived proteins associated with acute and chronic renal allograft rejection not connected to the network (not shown; n = 21). A2M: Alpha-2-macroglobulin; ACAN: Aggrecan core protein; ACTB: Actin, cytoplasmic 1; AGT: Angiotensinogen; AMBP: Alpha-1-microglobulin; APOA1: Apolipoprotein A1; APOA2: Apolipoprotein A-2; APOM: Apolipoprotein M; B2M: Beta-2-microglobulin; BCAN: Brevican core protein; CALR: Calreticulin-3; CD27: CD27 antigen; CFD: Complement factor D; COL1A1: Collagen alpha-1(I) chain; COL1A2: Collagen alpha-2(I) chain; COL3A1: Collagen alpha-1(III) chain; CTSZ: Cathepsin Z; DAG1: Dystroglycan; DEFA5: Defensin-5; DEFB1: β-defensin 1; DPP4: Dipeptidyl peptidase 4; EGF: Pro-epidermal growth factor; F2: Prothrombin; FABP4: Fatty acid-binding protein, adipocyte; FBXL19: F-box/LRR-repeat protein 19; FGA: Fibrinogen alpha chain; FGB: Fibrinogen beta chain; FGG: Fibrinogen gamma chain; FKBP1A: Peptidyl-prolyl cis-trans isomerase FKBP1A; HIST1H4B: Histone H4; HLA-DRB1: HLA-DRB1 protein; HP: Haptoglobin; HTRA1: Serine protease HTRA1; IGFBP7: Insulin-like growth factor-binding protein 7; IGHM: Ig mu chain C region; KITLG: Kit ligand; KNG1: Kininogen-1; KRT: Keratin, type II cytoskeletal; KRT9: Keratin, type I cytoskeletal 9; LBP: LPS-binding protein; LCAT: Phosphatidylcholine-sterol acyltransferase; LGALS3BP: Galectin-3-binding protein; LMAN2: Vesicular integral-membrane protein VIP36; LRG1: Leucine-rich alpha-2-glycoprotein; LUM: Lumican; PROS1: Vitamin K-dependent protein S; PSAP: Saposin B; SELL: L-selectin; SERPINA1: Alpha-1-antitrypsin; SERPINA10: Protein Z-dependent protease; SERPINA3: Alpha-1-anti-chymotrypsin; SERPINA5: Serine protease inhibitor; SERPINF1: Pigment epithelium-derived factor; SERPING1: Plasma protease C1 inhibitor; SHBG: Sex hormone-binding globulin; SUMO2: Small ubiquitin-related modifier 2; SUSD2: Sushi domain-containing protein 2; TIMP1: Metalloproteinase inhibitor 1; TTN: Titin; UMOD: Uromodulin; VGF: Neurosecretory protein VGF; CMA1: Chymase; CXCL10: C-X-C motif chemokine 10; DSTN: Destrin; ENAM: Enamelin; EVPL: Envoplakin; FN1: Fibronectin; GRB2: Growth factor receptor-bound protein 2; HNF1A: Hepatocyte nuclear factor 1-alpha; HPR: Haptoglobin-related protein; HSPA1A: Heat shock 70 kDa protein 1A; ITGA2: Integrin alpha-2; ITGA2B: Integrin alpha-IIb; ITGA5: Integrin alpha-5; ITGAM: Integrin alpha-M; ITGB1: Integrin beta-1; ITGB2: Integrin beta-2; ITGB3: Integrin beta-3; LGALS1: Galectin-1; MAX: Protein max; MMP8: Neutrophil collagenase; MYC: Myc proto-oncogene protein; MYOC: Myocilin; POLD1: DNA polymerase delta catalytic subunit; PRSS1: Trypsin-1; SERPINF2: Alpha-2-antiplasmin; SHC1: SHC-transforming protein 1; TAF1: Transcription initiation factor TFIID subunit 1; TANK: TRAF family member-associated NF-kappa-B activator; TGFBR1: TGF-beta receptor type-1; TNR: Tenascin-R; TRB@: T-cell receptor beta; TRIM63: E3 ubiquitin-protein ligase TRIM63; UGCGL1: UDP-glucose:glycoprotein glucosyltransferase 1; VCAN: Versican core protein; VIM: Vimentin; AFM: Afamin; CD5L: CD5 antigen-like; CLCA1: Calcium-activated chloride channel regulator 1; CLEC14A: C-type lectin domain family 14 member A; DPEP1: Dipeptidase; FAM151A: Protein FAM151A; FAM3C: Protein FAM3C; GGT6: Gamma-glutamyltransferase 6; GLB1: Beta-galactosidase; HAVCR2: Hepatitis A virus cellular receptor 2; KIAA0753: Uncharacterized protein KIAA0753; LGALS9B: Galectin-9B; MBL: Mannose-binding lectin; MMP-7: Matrilysin; MRC2: C-type mannose receptor 2; PGA4: Pepsin A-4; PI16: Peptidase inhibitor 16; RTN4RL2: Reticulon-4 receptor-like 2; SERPINA2P: Putative alpha-1-antitrypsin-related protein; SHISA5: Protein shisa-5; VASN: Vasorin.
- Citation: Gwinner W, Metzger J, Husi H, Marx D. Proteomics for rejection diagnosis in renal transplant patients: Where are we now? World J Transplant 2016; 6(1): 28-41
- URL: https://www.wjgnet.com/2220-3230/full/v6/i1/28.htm
- DOI: https://dx.doi.org/10.5500/wjt.v6.i1.28