Published online Jun 16, 2023. doi: 10.5497/wjp.v12.i3.25
Peer-review started: March 1, 2023
First decision: April 13, 2023
Revised: May 5, 2023
Accepted: May 31, 2023
Article in press: May 31, 2023
Published online: June 16, 2023
Processing time: 104 Days and 12.1 Hours
Some isopavines can exhibit important biological activity in the treatment of neurological disorders since it is considered an antagonist of the specific N-methyl-D-Aspartate (NMDA) receptor. Amurensinine is an isopavine which still has few studies. In view of the potential of isopavines as NMDA receptor antagonists, theoretical studies using bioinformatics were carried out in order to investigate whether Amurensinine binds to the NMDA receptor and to analyze the receptor/Ligand complex. This data can contribute to understanding of the onset of neurological diseases and contribute to the planning of drugs for the treatment of neurological diseases involving the NMDA receptor.
To investigate the interaction of the antagonist Amurensinine on the GluN1A/ GluN2B isoform of the NMDA receptor using bioinformatics.
The three-dimen-sional structure of the GluN1A/GluN2B NMDA receptor was selected from the Protein Data Bank (PDB) - PDB: 4PE5, and the three-dimen-sional structure of Amurensinine (ligand) was designed and optimized using ACD/SchemsketchTM software. Prediction of the protonation state of Amur-ensinine at physiological pH was performed using MarvinSketch software (ChemAxon). Protonated and non-protonated Amurensin were prepared using AutoDock Tools 4 software and simulations were performed using Autodock Vina v.1.2.0. The receptor/Ligand complexes were analyzed using PyMol (Schrödinger, Inc) and BIOVIA Discovery Studio (Dassault Systemes) software. To evaluate the NMDA receptor/Amurensinine complex and validate the molecular docking, simulations using NMDA receptor and Ifenprodil antagonist were performed under the same conditions. Ifenprodil was also designed, optimized and protonated, under the same conditions as Amurensinine.
Molecular docking simulations showed that both non-protonated and protonated Amurensinine bind to the amino terminal domain (ATD) domain of the GluN1A/GluN2B NMDA receptor with significant affinity energy, -7.9 Kcal/mol and -8.1 Kcal/mol, respectively. The NMDA receptor/non-protonated Amurensinine complex was stabilized by 15 bonds, while the NMDA receptor/protonated Amurensinine complex was stabilized by less than half, 6 bonds. Despite the difference in the number of bonds, the variation in bond length and the average bond length values are similar in both complexes. The complex formed by the NMDA receptor and Ifenprodil showed an affinity energy of -8.2 Kcal/mol, a value very close to that obtained for the NMDA receptor/Amurensinine complex. Molecular docking between Ifenprodil and the GluN1A /GluN2B NMDA receptor demonstrated that this antagonist interacts with the ATD of the receptor, which validates the simulations performed with Amurensinine.
Amurensinine binds to the NMDA receptor on ATD, similar to Ifenprodil, and the affinity energy is closer. These data suggest that Amurensinine could behave as a receptor inhibitor, indicating that this compound may have a potential biological application, which should be evaluated by in vitro and preclinical assays.
Core Tip: Amurensinine binds to a region of the amino terminal domain on the N-methyl-D-Aspartate receptor and the interaction is stabilized mainly by covalent bonds, which confer an affinity energy of significant value to the receptor/Ligand complex. The interaction between Amurensinine and the receptor, which is involved in neurological diseases, suggests that this isopavine may interfere with its function, so it may have therapeutic potential in this area.
- Citation: Façanha Wendel C, Hapuque Oliveira Alencar Q, Viana Vieira R, Teixeira KN. In silico insight into Amurensinine - an N-Methyl-D-Aspartate receptor antagonist. World J Pharmacol 2023; 12(3): 25-34
- URL: https://www.wjgnet.com/2220-3192/full/v12/i3/25.htm
- DOI: https://dx.doi.org/10.5497/wjp.v12.i3.25
Ion receptors are voltage-dependent ion channels which, when activated by an electrical potential difference, lead to the influx and/or efflux of ions. These receptors are classified into three main families: The N-Methyl-D-Aspartate (NMDA) receptors, the alpha-amino-3-hydroxy-methyl-5-4-isoxazolpropionic receptors and, the Kainate receptors[1].
Studies show that hyperactivation, inhibition or dysfunction of these receptors are related to the development of several diseases such as schizophrenia, stroke, Alzheimer disease[2], anti-NMDA receptor encephalitis[3] and depression[4]. Alterations in the NMDA receptor may also be associated with neural disorders such as epileptic aphasia and mental retardation[5,6].
During the resting potential, the NMDA receptor presents its subunits joined together[7-10] and its ion channel is blocked by Mg2+ ion. Depolarization of the postsynaptic membrane potential[1] removes the Mg2+ ion from the receptor channel entrance[11,12] and thus, the agonists Glycine and Glutamate are able to bind to the GluN1[13-16] and GluN2 subunits of the NMDA receptor, respectively[14,16].
The channel only returns to its normal state after the ligand binding domain (LBD) gap opens allowing the agonist to dissociate from the active site. The opening of the gap may be caused due to the binding of the agonist itself to the binding site; this triggers a conformational change that decreases the sensitivity of the active site[17-22].
Excitotoxicity is caused by excessive stimulation by neurotransmitters which can lead to cell damage or death. It may occur in events that characterize a central nervous system trauma, an ischemic or hemorrhagic condition, in which cells are deprived of energy to maintain ionic homeostasis[23]. The excess of Glutamate in the medium facilitates the activation of the NMDA receptor which allows the flux of Ca2+ ions into the cell. This intracellular Ca2+ accumulation can cause an osmotic swelling, lysis and cell death[23], activating enzymes such as proteases, phospholipases, and endonucleases, which can damage cell structures such as membranes and DNA itself.
In this situation, mitochondria are also harmed, because they are unable to buffer this excess of Ca2+, resulting in the formation of reactive oxygen species[23]. Neurons, which are positively charged (depolarized), promote the unblocking of the ion channel caused by Mg2+, facilitating a greater influx of ions through the channel, making it difficult to reestablish ionic homeostasis[24,25].
Allosteric modulators that bind to the amino terminal domain (ATD) are able to regulate the probability of receptor ion channel opening and its rate of closure, such as Zn2+ which binds to the GluN2A and GluN2B subunits, Ifenprodil that binds to GluN2B, and polyamines that bind to GluN2B[26-28]. LBD also has binding sites for agonist or antagonist allosteric modulators, capable of controlling ion channel opening[29,30].
Ifenprodil is a non-competitive antagonist that partially binds to the ATD of the GluN2B subunit of the NMDA receptor - where the Glutamate agonist binding site is located, and can inhibit its activity by up to 90% showing higher efficiency at GluN1/GluN2B compared to GluN1/GluN2A/GluN1/GluN2B[31-33].
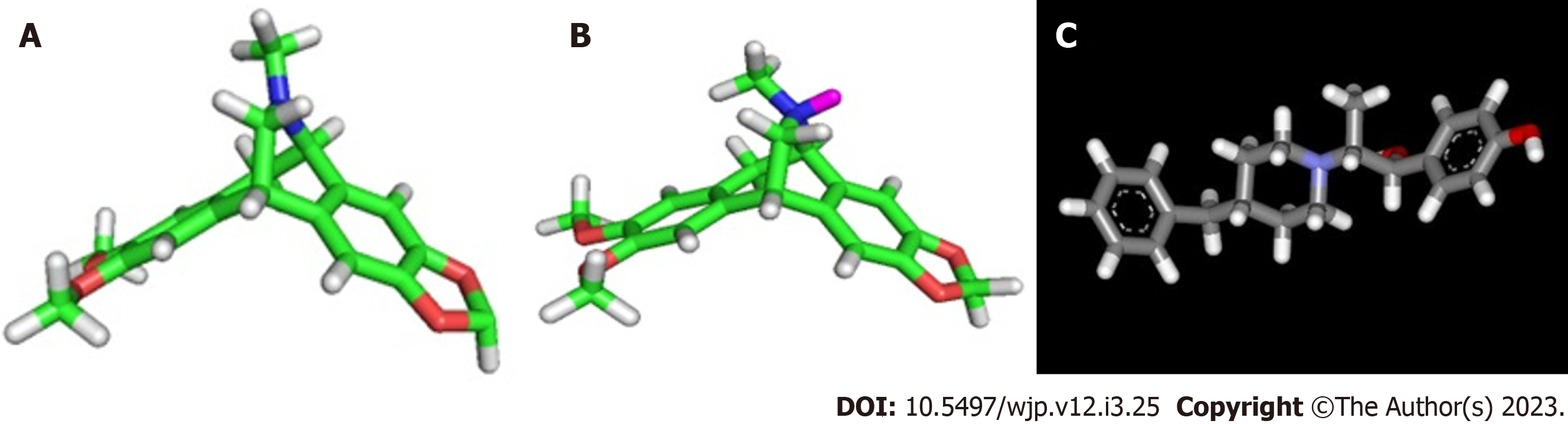
Isopavines are alkaloids derived from plants of the genus Papaver[34-37] and are classified as benzopyridine isoquinolines[37] that have in common the tetrahydroisoquinoline in the central region of their structures[38,39]. Isopavines are considered non-competitive and specific NMDA receptor ion channel antagonists or blockers[39]. Studies indicate that isopavines may exhibit important biological activity in the treatment of neurological disorders such as Down syndrome, Alzheimer, Huntington's disease, amyotrophic lateral sclerosis, senile dementia, stroke, epilepsy, and olivo-ponto-cerebellar atrophy[38,39]. Amurensinine is an isopavine that has been found in Meconopsis species[40] and it has been identified in Papaver alpinum, Papaver tatricum, Papaver pyrenaicum, Papaver suaveolens and some varieties of Papaver nudicaule[34-37].
Molecular docking is a computational method used to predict possible sites of interaction of a ligand in a receptor, as well as the affinity of the interaction between receptor-ligand, the conformation of the receptor-ligand complex and the nature of the chemical bonds between the receptor and the ligand in order to define the stability of the interaction. This method has great importance for the development of new drugs, because it allows you to optimize the design of a drug, to find a candidate drug by virtual scanning in databases[41].
Therefore, molecular docking simulations between Amurensinine and the NMDA receptor were performed in order to evaluate the receptor/Ligand complex, and thereby provide data to support future research on neurological diseases involving this receptor.
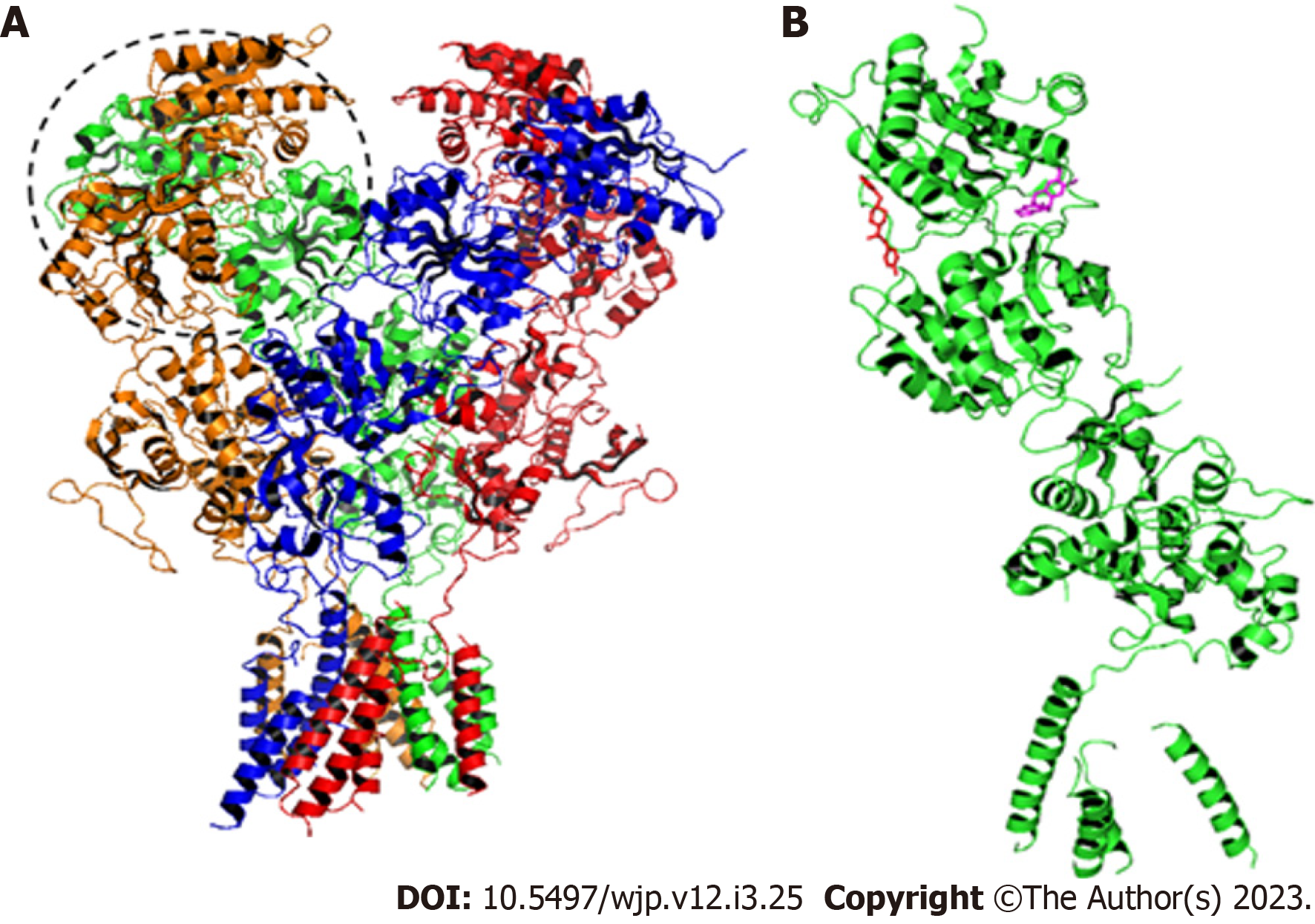
This study was performed with the GluN1A/GluN2B isoform of the NMDA receptor. The native mutation-free three-dimensional structure of the human GluN1A/GluN2B NMDA receptor is not available on the structure databases, thus the structure of the receptor from a mammal used in preclinical trials - Rattus norvegicus - was selected. The three-dimensional structure of the GluN1A/GluN2B NMDA receptor was selected from the Protein Data Bank (PDB) (www.rcsb.org) - PDB 4PE5. Three-dimensional structure of Amurensinine (ligand) was designed and optimized using ACD/SchemsketchTM software. The protonation state prediction of the Amurensinine at physiological pH 7.4 was performed using the MarvinSketch software (ChemAxon). Molecular docking simulations were performed with Amurensinine in its protonated and non-protonated states.
Molecular docking was performed using the flexible ligand - rigid receptor methodology[42]. Simulations were performed with the B-chain of the GluN2B subunit of the GluN1A/GluN2B NMDA receptor, where the ATD - domain in which the binding site of the antagonist Ifenprodil, is located in human receptor. For the execution of molecular docking, the AutoDock Tools 4[43] and Autodock Vina v.1.2.0.[44] software was used. In these software the steps for the preparation of molecules were performed, the torsion points of ligand were detected, its torsion angles were calculated and Grid box dimensions were determined and the command was run by AutoDock Vina. Data were analyzed using PyMol (Schrödinger, Inc) and BIOVIA Discovery Studio (Dassault Systemes) software.
To validate and analyze the NMDA receptor/Amurensinine complex, the molecular docking was performed with the Ifenprodil antagonist (Figure 1 under the same conditions. Ifenprodil was also designed, optimized and protonated as well as Amurensinine. Ifenprodil was docked on the ATD of the GluN1A/GluN2B NMDA receptor (PDB 4PE5).
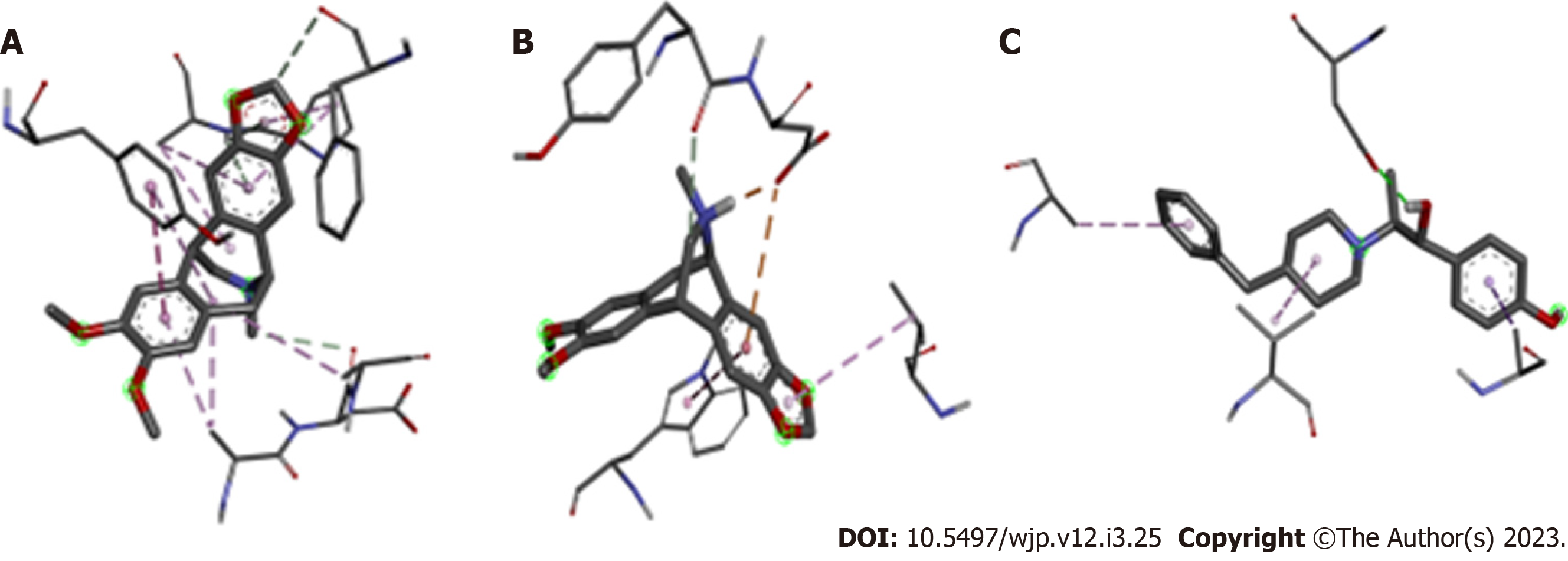
At physiological pH (pH = 7.4) it was observed that 68.5% of Amurensinine was in its non-protonated state and 31.5% was in the protonated one; both states show structural differences (Figure 1). Molecular docking showed that Amurensinine binds to the NMDA receptor; both protonated and non-protonated Amurensinine binds to the ATD of the GluN1A/GluN2B NMDA receptor with significant affinity energy, -7.9 Kcal/mol and -8.1 Kcal/mol, respectively. Despite the similar affinity energy values, the non-protonated Amurensinine makes 15 bonds with regions of the ATD, while the protonated compound makes only six bonds (Table 1). However, the variation in bond lengths and the average of these lengths in both the non-protonated Amurensinine/NMDA receptor complex (2.9-5.4 Å; average = 4.23) and the protonated Amurensinine/NMDA receptor complex (2.0-5.7 Å; average = 3.90) are similar. Upon coupling of Ifenprodil to ATD, an energy affinity = -8.2 Kcal/mol was obtained, a value very close to that obtained for protonated Amurensinine/GluN1A/GluN2B NMDA receptor complex; even the same number of bonds was observed (six bonds) (2.6-4.9 Å; average = 3.67) as shown in Table 2; however, the geometry of the bonds is different (Figure 2). This result is consistent, since the structures of Amurensinine and Ifenprodil are similar, but not identical (Figure 3). The interaction between Ifenprodil and ATD on NMDA receptor validates the molecular docking of Amurensinine and confers reliability to the data.
| Non protonated Amurensinine: Receptor1 | Distance (Å) | Bond type |
| Aromatic ring (A): ARG347 | 4.1 | π-alky1 |
| Aromatic ring (A): TYR 287 | 4.8 | π-π T-shaped |
| Aromatic ring (B): LYS361 | 2.9 | Hydrogen bond |
| Aromatic ring (B): LYS361 | 4.8 | π-alky1 |
| Aromatic ring (B): PRO360 | 5.4 | π-alky1 |
| C-12: ARG347 | 4.2 | Alky1 |
| C-12: LEU349 | 5.2 | Alky1 |
| C-12: TYR287 | 5.1 | π-alky1 |
| C-22: ASP348 | 3.6 | Carbon-hydrogen bond |
| C-9: PHE146 | 3.4 | Carbon-hydrogen bond |
| Cycloheptane: LYS361 | 4.8 | Alky1 |
| Cyclopentane: LYS361 | 3.3 | Hydrogen bond |
| Cyclopentane: PRO360 | 5 | π-alky1 |
| N-1: HIS359 | 3.3 | Hydrogen bond |
| O-3: ASP295 | 3.5 | Hydrogen bond |
| Protonated Amurensinine: Receptor* | Distance (Å) | Bond type |
| Aromatic ring (B): ASP477 | 4.4 | π-anion |
| Aromatic ring (B): TRP166 | 5.7 | Alky1 |
| CH-19: TYR476 | 3.1 | Carbon-hydrogen bond |
| Cyclopentane: VAL390 | 4.9 | π- alky1 |
| NH-1: ASP477 | 2 | Carbon-hydrogen bond |
| O1: ASP165 | 3.3 | Hydrogen bond |
Neurological diseases are an important cause of morbidity and mortality and loss of quality of life. Their pathophysiology is complex and multifactorial, since it involves genetic and environmental factors, resulting in different clinical manifestations in patients with the same neurological damage[45].
Elucidated mechanisms involved in the onset of neuronal diseases include hyperactivation, inhibition and dysfunction of the NMDA receptor, which is involved in schizophrenia, stroke, Alzheimer[2], anti-NMDAR encephalitis[3], depression[4], mental retardation and epileptic aphasia[5,6]. This receptor is an ion channel composed of four subunits and has three families - GluN1, GluN2 (A-D isoforms) and GluN3 (A-B isoforms)[46]. Each receptor subunit is composed of two extracellular domains - LBD and ATD, an intracellular carboxy terminal domain (CTD) and a transmembrane domain (TMD), where the ion channel is located[7,16,39,47-49].
Therefore, compounds which regulate the opening and closing speed of the ion channel by binding to ATD, are promising for research into the treatment of neurological diseases[39,49]. In silico, the Amurensinine binds to the ATD of the GluN1A/GluN2B NMDA receptor with significant affinity. The affinity is measured by affinity energy which, when it is lower than -6.0 kcal/mol, indicates interaction in a biological environment[50]. This data is important once it indicates the possibility of interaction of the Amurensinine with the target receptor inside the organism. Ifenprodil is known to bind to the GluN2B subunit of the human NMDA receptor[32]; by binding in the same region of the NMDA receptor from Rattus norvegicus as Amurensinine, it is suggested that this isopavine may also interact with the human receptor. In addition, the low value of affinity energy points out the stability of the Amurensinine/NMDA receptor complex, and consequently indicates chances of pharmacological effectiveness. However, these assumptions need to be tested experimentally, in vitro and in vivo.
NMDA receptors normally function at physiological pH - neutral[44]; in cases of brain damage or neural disease, this pH may decrease and the environment becomes acidic[44,45]. The interaction of Amurensinine with the NMDA receptor in both protonated and non-protonated states, with similar affinities, indicate that the pH and, consequently, the protonation state of this compound, do not influence its interaction. Thus, neurological disorders that alter the H+ concentration in the environment will not have a significant influence on the modulating activity of Amurensinine on the receptor.
An opposite result was observed with Endobain E and the NMDA receptor. This compound is an endogenous brain factor that acts as an inhibitor of Na/K ATPase and, as a modulator of the NMDA receptor by binding to the inner surface of the channel, decreasing the affinity of receptor ligands. Under conditions of cerebral ischemia, Endobain E has its activity optimized due to the acidic environment of the ischemic brain (pH 6.5, 90% receptor inhibition). In a more alkaline environment, its activity is reduced (pH 7.4-8, 25% receptor inhibition)[51].
In general, proteins such as NMDA receptor-forming ones, have their structural conformation and, consequently, their activity directly influenced by pH. The sudden change in H+ concentration induces ionization of the residues, conferring an excessively negative or positive charge, which can lead to intramolecular repulsion, exposure of hydrophobic regions, and loss of function[52]. On the other hand, pH variation can also expose previously hidden sites, optimizing the receptor-ligand interaction, as occurs with Endobain E. Regarding Amurensinine, it is inferred that its binding site is not in a region of the receptor that undergo major conformational changes with pH variation of the medium and that this site is capable of binding both the protonated and non-protonated states of this isopavine.
The binding strength, also called intermolecular interaction, occurs between covalently interacting compounds and determines the effectiveness of the receptor-ligand interaction[53]. Moreover, the bond type that molecules establish with each other directly influences their physicochemical properties, such as solubility and boiling point, and their final conformation. In enzymes the type of contact established is able to alter their activity, strength, selectivity and the conformation of the enzyme-substrate complex[54]. Thus, predicting the strength and type of interaction between receptor and ligand is important to understand the mechanism of action of the ligand, which may be a drug candidate.
Amurensinine in protonated state interacts with the receptor through six chemical bonds of which the majority (83%) are covalent bonds. Amurensinine in the non-protonated state made 15 chemical bonds with the receptor, with a predominance of covalent bonds, and even with the large difference in the number of bonds, the affinity energy was quite similar for both receptor/Ligand complexes. Possibly, a contributing factor to the similarity of affinities is the shorter bond lengths that stabilize the protonated Amurensinine/receptor complex.
In this regard, Medeiros[55] cites the physicochemical characteristics of the interaction of antagonistic compounds, such as Ifenprodil, with the ATD in the GluN2B subunit of the NMDA receptor. This compound, despite having a strong interaction with the receptor, has little selectivity as it also binds to the Alpha-1 and 5HT receptors, leading to impairments in motor function and a drop in blood pressure. However, its use is of fundamental importance as a parameter for studying compounds that interact differently with the binding site, such as MK-22, EVT-101, and Amurensinine[55].
Amurensinine, in both protonated and non-protonated states, binds to the ATD region of the GluN2B subunit of the murine GluN1A/GluN2B NMDA receptor, in silico, forming complexes stabilized mainly by covalent bonds. The affinity energy of the complexes is significantly low and indicates, in addition to probability of interaction in the physiological environment, the stability of the complexes. The structural and receptor interaction similarity between Amurensinine and Ifenprodil suggest that this isopavine could behave as a receptor inhibitor; therefore, this compound could present potential biological application, which needs to be evaluated by in vitro and in vivo assays.
Isopavines are alkaloids derived from plants of the genus Papaver that have biological activity for the treatment of neurological disorders. Amurensinine is a little-studied isopavine whose activity in relation to neurological disorders has not been studied. Research on this isopavine may contribute new information about its action.
The research motivation was the scarce literature on the subject. There are few studies in health care using isopavines; similarly, studies on Amurensinine are quite scarce. Therefore, we studied Amurensinine in silico to verify its preliminary therapeutic potential.
To study, in silico, the interaction between Amurensinine and the N-methyl-D-Aspartate (NMDA) receptor, which is involved in the onset of neurological disorders.
In this study we used molecular docking, a standardized bioinformatics methodology for analyzing chemical interactions between receptors and ligands.
The research results indicated that Amurensinine can interact with the NMDA receptor with high affinity, and that its protonation state does not significantly interfere with this affinity.
The results of the research were satisfactory, as Amurensinine was able to bind to the tNMDA receptor. The occurrence of interaction indicates that this isopavine may interfere with the activity of the receptor. Since it binds with similar affinity and to the same subunit as the antagonist Ifenprodil, it may act as an inhibitor of the receptor. However, in vitro and in vivo studies are needed to affirm this.
The research perspective is to conduct further in silico analyses with Amurensinine and other receptors involved in the onset of neurological diseases to further evaluate the potential of this isopavine for health care.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Pharmacology and pharmacy
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Chen X, United States; Emran TB, Bangladesh S-Editor: Li L L-Editor: A P-Editor: Ji MX
| 1. | Carcia P. Perpendicular magnetic anisotropy in Pd/Co and Pt/Co thin‐film layered structures. J Appl Phys. 1988;63: 5066-5073. [DOI] [Full Text] |
| 2. | Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1443] [Cited by in RCA: 1868] [Article Influence: 155.7] [Reference Citation Analysis (0)] |
| 3. | Dalmau J, Gleichman AJ, Hughes EG, Rossi JE, Peng X, Lai M, Dessain SK, Rosenfeld MR, Balice-Gordon R, Lynch DR. Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. Lancet Neurol. 2008;7:1091-1098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2443] [Cited by in RCA: 2154] [Article Influence: 126.7] [Reference Citation Analysis (0)] |
| 4. | Moskal JR, Burch R, Burgdorf JS, Kroes RA, Stanton PK, Disterhoft JF, Leander JD. GLYX-13, an NMDA receptor glycine site functional partial agonist enhances cognition and produces antidepressant effects without the psychotomimetic side effects of NMDA receptor antagonists. Expert Opin Investig Drugs. 2014;23:243-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 5. | Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, Milh M, Kortüm F, Fritsch A, Pientka FK, Hellenbroich Y, Kalscheuer VM, Kohlhase J, Moog U, Rappold G, Rauch A, Ropers HH, von Spiczak S, Tönnies H, Villeneuve N, Villard L, Zabel B, Zenker M, Laube B, Reis A, Wieczorek D, Van Maldergem L, Kutsche K. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 400] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 6. | Lesca G, Rudolf G, Bruneau N, Lozovaya N, Labalme A, Boutry-Kryza N, Salmi M, Tsintsadze T, Addis L, Motte J, Wright S, Tsintsadze V, Michel A, Doummar D, Lascelles K, Strug L, Waters P, de Bellescize J, Vrielynck P, de Saint Martin A, Ville D, Ryvlin P, Arzimanoglou A, Hirsch E, Vincent A, Pal D, Burnashev N, Sanlaville D, Szepetowski P. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 318] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 7. | Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1852] [Cited by in RCA: 1948] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 8. | Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 153] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319-321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 628] [Cited by in RCA: 546] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 10. | Ulbrich MH, Isacoff EY. Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci U S A. 2008;105:14163-14168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 131] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 11. | Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1992] [Cited by in RCA: 2086] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 12. | Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2796] [Cited by in RCA: 2935] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 13. | Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873-2885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 387] [Cited by in RCA: 385] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 14. | Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 567] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 15. | Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 2008;27:2158-2170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 16. | Lü W, Du J, Goehring A, Gouaux E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science. 2017;355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 138] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 17. | Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 548] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 18. | Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 301] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 19. | Jin L, Sugiyama H, Takigawa M, Katagiri D, Tomitori H, Nishimura K, Kaur N, Phanstiel O 4th, Kitajima M, Takayama H, Okawara T, Williams K, Kashiwagi K, Igarashi K. Comparative studies of anthraquinone- and anthracene-tetraamines as blockers of N-methyl-D-aspartate receptors. J Pharmacol Exp Ther. 2007;320:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Horning MS, Mayer ML. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 176] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 22. | Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol. 2006;13:1120-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 92] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Lipton SA, Nicotera P. Calcium, free radicals and excitotoxins in neuronal apoptosis. Cell Calcium. 1998;23:165-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 137] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 24. | Zeevalk GD, Nicklas WJ. Evidence that the loss of the voltage-dependent Mg2+ block at the N-methyl-D-aspartate receptor underlies receptor activation during inhibition of neuronal metabolism. J Neurochem. 1992;59:1211-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 96] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Kushnareva YE, Wiley SE, Ward MW, Andreyev AY, Murphy AN. Excitotoxic injury to mitochondria isolated from cultured neurons. J Biol Chem. 2005;280:28894-28902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Yang HX, Chshiev M, Dieny B, Lee JH, Manchon A, Shin KH. First principles investigation of the very large perpendicular magnetic anisotropy at Fe|MgO and Co|MgO interfaces. Phys Rev. 2011;84:054401-054405. [DOI] [Full Text] |
| 27. | Cubukcu M, Boulle O, Drouard M, Garello K, Avci CO, Miron IM, Langer J, Ocker B, Gambardella P, Gaudin G. Spin-orbit torque magnetization switching of a three-terminal perpendicular magnetic tunnel junction. Appl Phys Lett. 2014;104:042406. [DOI] [Full Text] |
| 28. | Ikeda S, Sato H, Yamanouchi M, Gan H, Miura K, Mizunuma K, Kanai S, Fukami S, Matsukura F, Kasai N, Ohno H. Recent progress or perpendicular anisotropy magnetic tunnel junctions for nonvolatile VLSI. SPIN. 2012;2:1240003. [DOI] [Full Text] |
| 29. | Gajek M, Nowak JJ, Sun JZ, Trouilloud PL, O'Sullivan EJ, Abraham DW, Gaidis MC, Hu G, Brown S, Zhu Y, Robertazzi RP, Gallagher WJ, Worledge DC. Spin torque switching of 20 nm magnetic tunnel junctions with perpendicular anisotropy. Appl Phys Lett. 2012;100:132408. [DOI] [Full Text] |
| 30. | Zhu S, Paoletti P. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr Opin Pharmacol. 2015;20:14-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 132] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 31. | Williams K. Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: selectivity and mechanisms at recombinant heteromeric receptors. Mol Pharmacol. 1993;44:851-859. |
| 32. | Hess SD, Daggett LP, Deal C, Lu CC, Johnson EC, Veliçelebi G. Functional characterization of human N-methyl-D-aspartate subtype 1A/2D receptors. J Neurochem. 1998;70:1269-1279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Hatton CJ, Paoletti P. Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron. 2005;46:261-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 190] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 34. | Boit H-G, Flentje H. Neue Alkaloide aus Papaver amurense. Naturwiss. 1959;46:514-515. [RCA] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 35. | Boit H-G, Flentje H. Nudaurin, Muramin und Amurensin, drei neue Papaver-Alkloide. Naturwiss. 1960;47:180. [RCA] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 18] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 36. | Maturová M, Moza BK, Sitar J, Santavy F. Isolierung der alkaloide aus einigen papaverarten der sektion scapiflora reichb. - isolierung der alkaloide aus einigen papaverarten vi1. Planta Medica. 1962;10:124. [DOI] [Full Text] |
| 37. | Maturová M, Pavlásková D, Santavý F. [Isolation of alkaloids from some types of the papaver genus. Isolation and chemistry of alkaloids of some types of papaver. XXXIV]. Planta Med. 1966;14:22-41. [PubMed] [DOI] [Full Text] |
| 38. | Weber E, Keana J, Barmettler P. The PCT receptor ligands and the use thereof. State of Oregon patent, acting by and through the Oregon State Board of Higher Education, on behalf of Oregon Health Science University and the University of Oregon AU643205B2. Apr 13, 1990. [cited 23 May 2023]. Available from: https://patents.justia.com/assignee/state-of-oregon-acting-by-and-through-the-oregon-state-board-of-higher-education-on-behalf-of-the-oregon-health-sciences-university-a-non-profit-organization. |
| 39. | Wayne E, Childers JR, Magid A, Abou-Gharbia G 10, 11-dihydro-5-alkyl-12-substituted-10, 5-(minomethano)-5hdibenzoadcycloheptenes as neuroprotectant agents. American Home Products Corporation patent U.S. US4940789A. Jul 10, 1990. [cited 23 May 2023]. Available from: https://patents.google.com/patent/US4940789A. |
| 40. | Slavfk J; SLAVIKOVA, L. Alkaloids of some himalayan species of Meconopsis genus. Chem Commun. 1977;42:132-139. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 41. | Morris GM, Lim-Wilby M. Molecular docking. Methods Mol Biol. 2008;443:365-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 479] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 42. | Lengauer T, Rarey M. Computational methods for biomolecular docking. Curr Opin Struct Biol. 1996;6:402-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 415] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 43. | Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785-2791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19127] [Cited by in RCA: 16158] [Article Influence: 1009.9] [Reference Citation Analysis (0)] |
| 44. | Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J Chem Inf Model. 2021;61:3891-3898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1065] [Cited by in RCA: 2666] [Article Influence: 666.5] [Reference Citation Analysis (0)] |
| 45. | Dumurgier J, Tzourio C. Epidemiology of neurological diseases in older adults. Rev Neurol (Paris). 2020;176:642-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 46. | Hashimoto K. The NMDA Receptors. 1th ed. Cham: Springer International Publishing, 2017. [DOI] [Full Text] |
| 47. | Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1337] [Cited by in RCA: 1373] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 48. | Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405-496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2956] [Cited by in RCA: 2727] [Article Influence: 181.8] [Reference Citation Analysis (0)] |
| 49. | Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 484] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 50. | Pantsar T, Poso A. Binding Affinity via Docking: Fact and Fiction. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 316] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 51. | Reinés A, Zárate S, Carmona C, Negri G, Peña C, Rodríguez de Lores Arnaiz G. Endobain E, a brain endogenous factor, is present and modulates NMDA receptor in ischemic conditions. Life Sci. 2005;78:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 52. | Marzzoco A. Bioquímica Básica. 4th ed. Guanabara Koogan LTDA, 2015. |
| 53. | House J. Consumer acceptance of insect-based foods in the Netherlands: Academic and commercial implications. Appetite. 2016;107:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 152] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 54. | Cserháti T, Szögyi M. Role of hydrophobic and hydrophilic forces in peptide-protein interaction: new advances. Peptides. 1995;16:165-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 55. | Medeiros NVP. Estudos in silico de inibidores alostéricos do domínio amino terminal de receptores NMDA que contenham a subunidade GLUN2B. Master's thesis, Universidade Federal do Rio Grande do Norte. 2021. [cited 23 May 2023]. Available from: https://repositorio.ufrn.br/bitstream/123456789/46842/1/Estudossilicoinibidores_Medeiros_2022.pdf. |