Copyright
©The Author(s) 2023.
World J Pharmacol. Jun 16, 2023; 12(3): 25-34
Published online Jun 16, 2023. doi: 10.5497/wjp.v12.i3.25
Published online Jun 16, 2023. doi: 10.5497/wjp.v12.i3.25
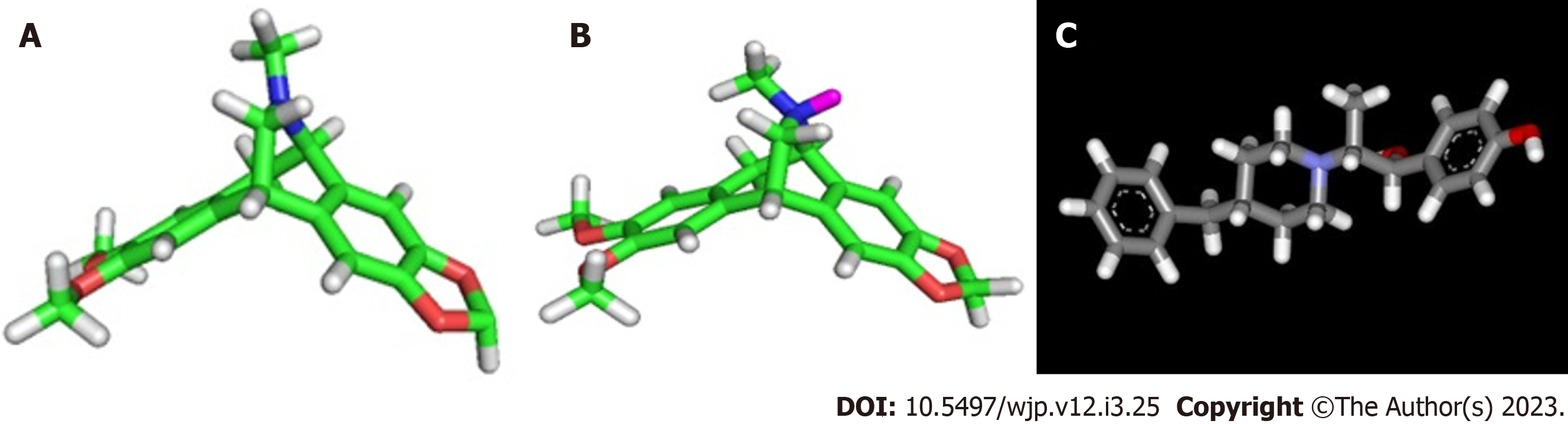
Figure 1 Structures of Amurensinine and Ifenprodil.
A: Amurensinine in the non-protonated state; B: Amurensinine in the protonated state at pH 7.4; C: Ifenprodil. These structures were designed using ACD/Schemsketch software.
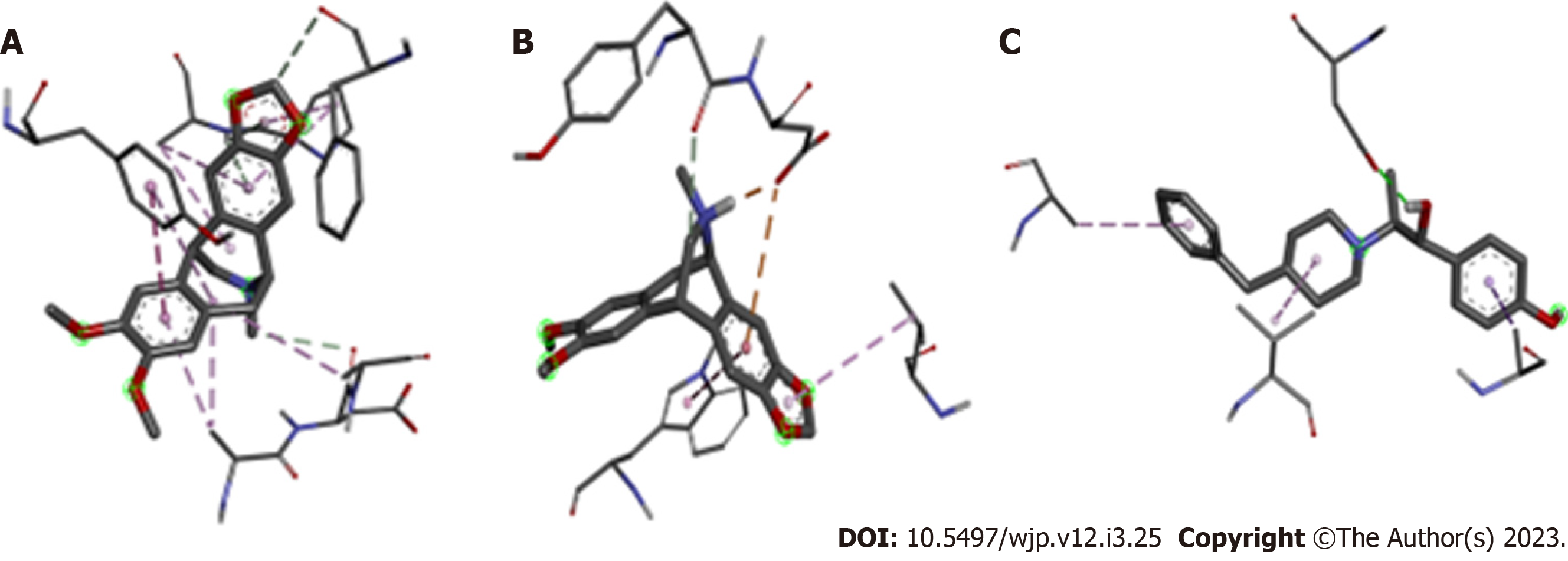
Figure 2 Binding geometry of antagonists to the amino terminal domain on the GluN1A/GluN2B N-methyl-D-Aspartate receptor.
A: Non-protonated Amurensinine; B: Protonated Amurensinine; C: Ifenprodil. The structures in thick sticks are the antagonists; the thin sticks are the amino acid residues of the receptor. Chemical bonds are represented by dashed lines. These structures were designed using ACD/Schemsketch software.
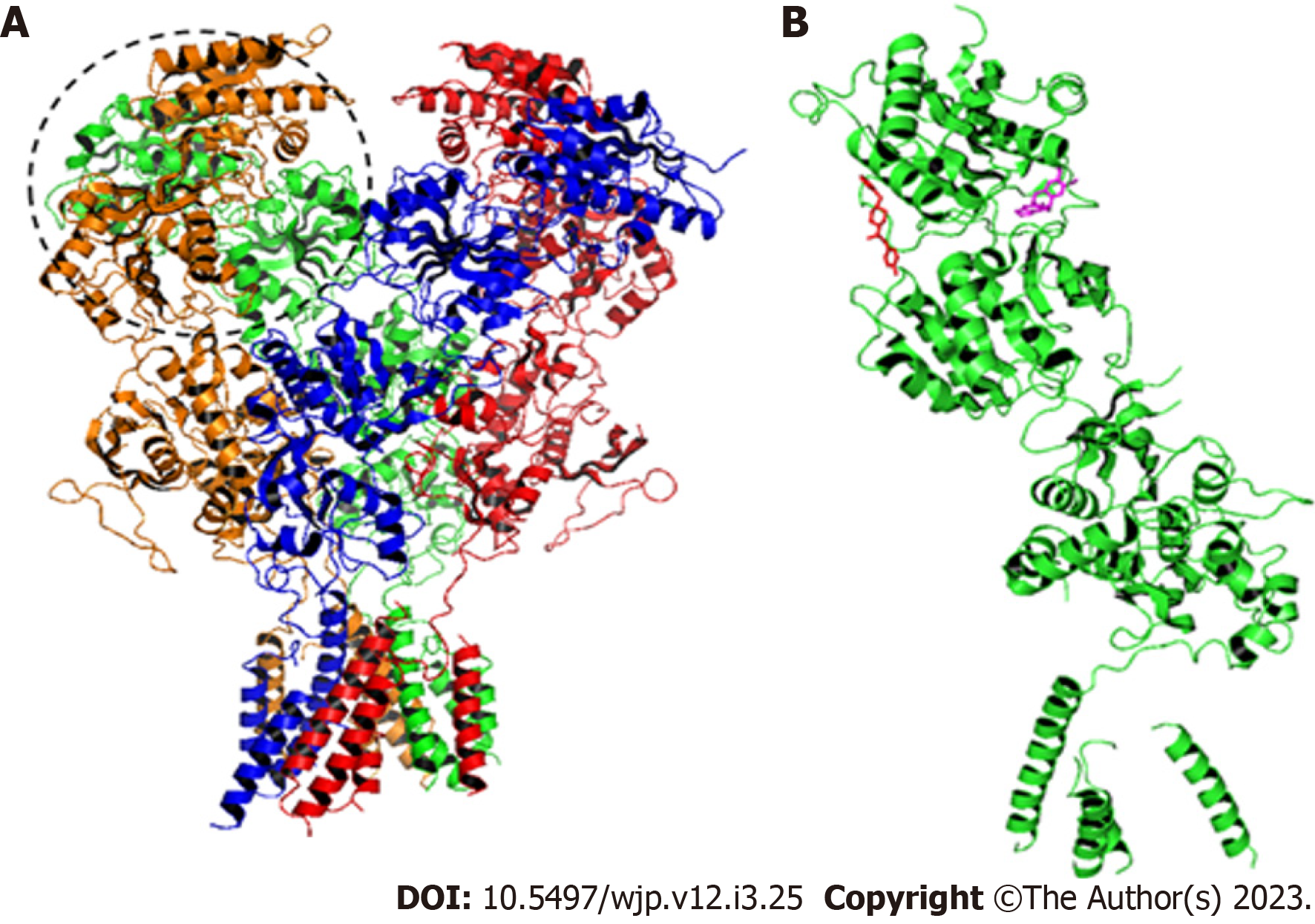
Figure 3 Structure of the GluN1A/GluN2B N-methyl-D-Aspartate receptor.
A: The dashed circle indicates the amino terminal domain (ATD). A-Chain: Orange; B-Chain: Green; C-Chain: Red; D-Chain: Blue. In the B-Chain are located the binding sites for Amurensinine and Ifenprodil; B: B-Chain with Ifenprodil (red) and non-protonated Amurensinine (pink) bound to the ATD. These structures were designed using ACD/Schemsketch software.
- Citation: Façanha Wendel C, Hapuque Oliveira Alencar Q, Viana Vieira R, Teixeira KN. In silico insight into Amurensinine - an N-Methyl-D-Aspartate receptor antagonist. World J Pharmacol 2023; 12(3): 25-34
- URL: https://www.wjgnet.com/2220-3192/full/v12/i3/25.htm
- DOI: https://dx.doi.org/10.5497/wjp.v12.i3.25