Copyright
©2014 Baishideng Publishing Group Inc.
World J Med Genet. May 27, 2014; 4(2): 27-33
Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.27
Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.27
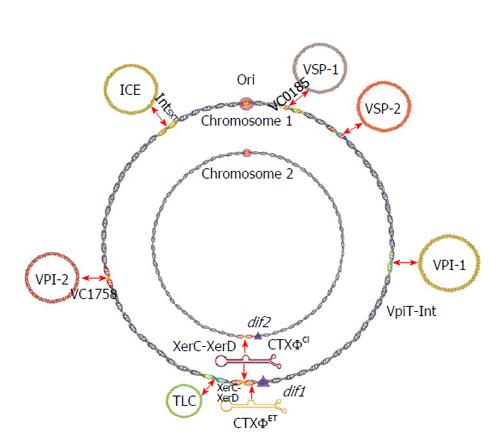
Figure 1 Schematic representation of the integrative mobile genetic elements present in the chromosome 1 (large) or 2 (small) of Vibrio cholerae.
Except CTXΦ, all other IMGEs have unique attB site either in large or small chromosome. IMGEs present in the dif region exploit host-encoded recombinases for integration while the genome of other IMGEs encode their own recombinases for integration (see detail in the text). Except CTXΦ, integration of rest of the IMGEs relies on their dsDNA genome and the integration is reversible. IMGE: Integrative mobile genetic element; CTXΦ: Cholera toxin phage; ICE: Integrative conjugative elements; VPI: Vibrio pathogenicity island; VSP: Vibrio seventh pandemic; TLC: Plasmids; dif: Dimer resolution sites.
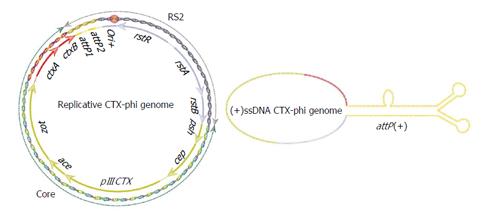
Figure 2 Replicative and integrative genomes of cholera toxin phage.
Replicative genome of CTXΦ arranged in structurally and functionally distinct two modular structures RS2 and core. RS2 encoded proteins are essential for phage replication, integration and transcriptional regulation of phage genes. Core encoded proteins are essential for phage morphogenesis and virion production. Folded (+)ssDNA phage genome is essential for phage integration. Functional XerCD binding site, attP(+), is formed by complementary base pairing between attP1 and attP2 of (+)ssDNA of phage genome. We have used similar color codes for replicative and integrative phage genomes for easy understanding of genetic attributes of attP(+) region. CTXΦ: Cholera toxin phage; RS2: Repeat sequence 2.
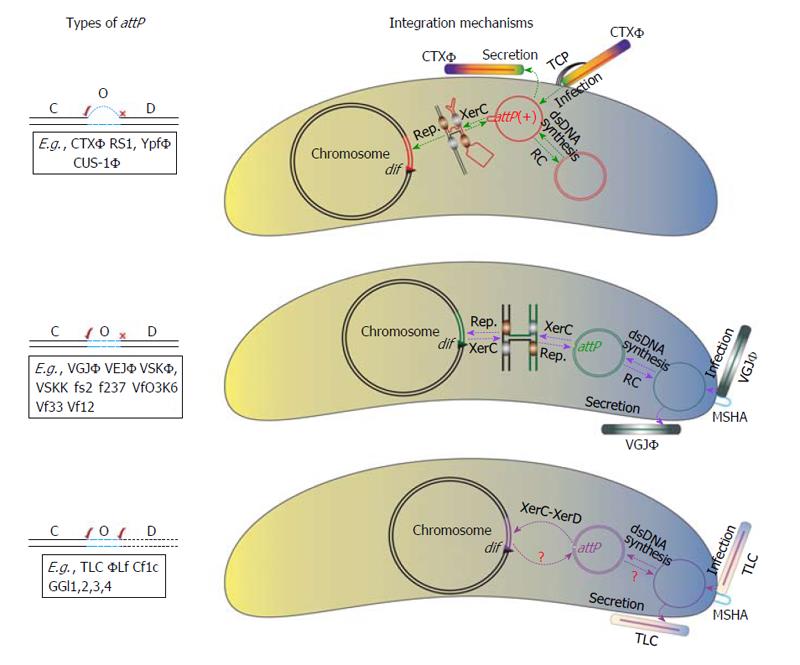
Figure 3 Schematic representation of the integration mechanisms of three classes of integrative mobile genetic elements present exclusively in the dimer resolution sites of bacterial genome.
C-O-D denotes XerC-Overlap region-XerD. The XerC-XerD recombinases, attachment site of phage and bacterial genome are essential components of nucleoprotein complex. Compatibility of terminal bases attP with attB immediate after XerC or XerD cleavage is indicated by √. The nucleoprotein complex and the sequential strand exchanges are not yet characterized for TLC and related genetic elements. CTXΦ integration is irreversible. By contrast, VGJΦ and TLC integration is reversible. Key steps in the life cycle of CTXΦ, VGJΦ and TLC are also indicated. Host-encoded TCP and MSHA served as phage receptors for CTXΦ, VGJΦ and TLC, respectively. Generally, TLC exploit fs2 encoded protein for encapsulation of its ssDNA genome and produces virion. Virion recognizes specific receptor present on its host cell surfaces and delivers its single-stranded DNA genome into the host cytoplasm. Once inside the host cell, the ssDNA phage genome is either converted into a double-stranded replicative phage genome by host machineries or directly integrated into the host chromosome. TLC: Plasmids; CTXΦ: Cholera toxin phage; TCP: Toxin co-regulated pilus; MSHA: Mannose sensitive hemagglutinin A.
-
Citation: Das B, Nair GB, Bhadra RK. Acquisition and dissemination mechanisms of CTXΦ in
Vibrio cholerae : New paradigm fordif residents. World J Med Genet 2014; 4(2): 27-33 - URL: https://www.wjgnet.com/2220-3184/full/v4/i2/27.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v4.i2.27