Copyright
©2014 Baishideng Publishing Group Inc.
World J Clin Infect Dis. Nov 25, 2014; 4(4): 41-49
Published online Nov 25, 2014. doi: 10.5495/wjcid.v4.i4.41
Published online Nov 25, 2014. doi: 10.5495/wjcid.v4.i4.41
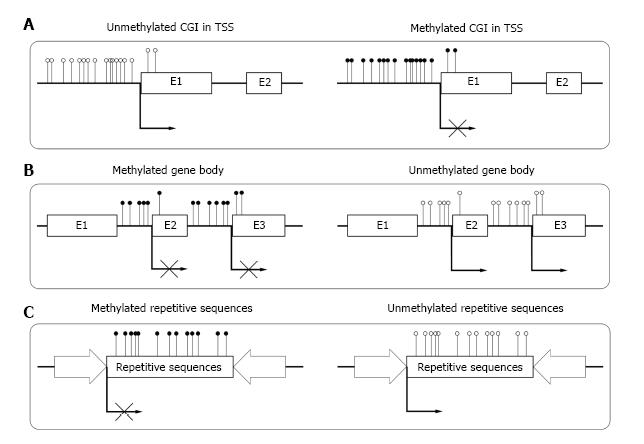
Figure 1 DNA methylation pattern in different parts of the genomes.
The normal conditions are presented in the left column and aberrant conditions are shown on the right. The black dots represent methylated CpG sites and the white circles represent unmethylated CpG sites. A: In normal cells, CpG islands (CGI) in transcriptional start site (TSS) usually remain unmethylated, allowing transcription. Aberrant methylation often links to long-term stabilization of transcriptional silencing and loss of gene function both physically and pathologically; B: In normal cells, gene bodies are CpG-poor and extensively methylated, increasing elongation efficacy. Aberrant demethylation of gene bodies may facilitates spurious initiations of transcription; C: In normal cells, repetitive sequences of genome are highly methylated, preventing chromosomal instability or gene disruption. Aberrant demethylation of repetitive sequences may reactivate endoparasitic sequences.
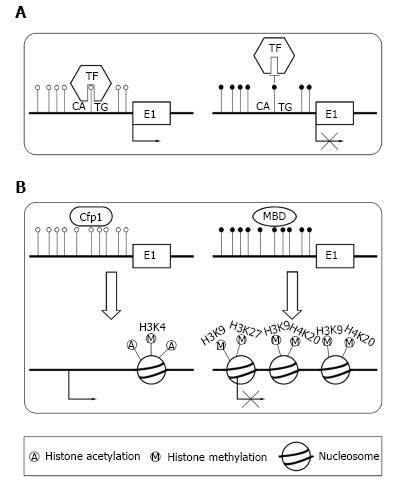
Figure 2 Transcriptional suppression mechanisms of DNA methylation in TSSs.
The normal conditions are presented in the left column and aberrant conditions are shown on the right. The black dots represent methylated CpG sites and the white circles represent unmethylated CpG sites. A: In normal cells, transcription factors (TF) bind to unmethylated binding site, allowing transcription. Aberrant methylated binding site prevent TF binding to its normal sites; B: In normal cells, unmethylated CpG island can recruit CpG binding proteins (Cfp1) and trigger histone modifications characterized by high levels of acetylation and trimethylated H3K4, H3K36 and H3K79. Finally, it forms a structure suitable for transcription. Aberrant methylated recruit methyl-CpG-binding domain (MBD) proteins and trigger histone modifications characterized by high levels of H3K9, H3K27 and H4K20 methylation and low levels of acetylation. It represses the transcriptional permissiveness of chromatins and results in gene silencing.
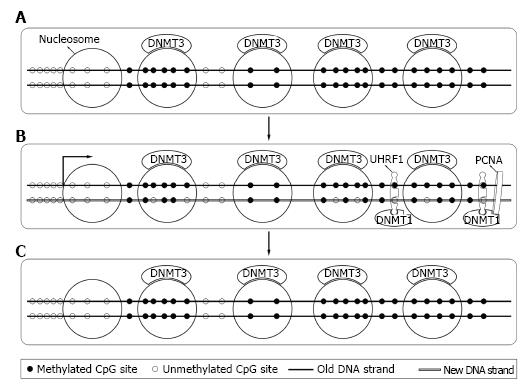
Figure 3 The maintenance of DNA methylation pattern.
A: In somatic cells, DNA methyltransferase (DNMT) 3 (DNMT 3a and DNMT 3b) are bound to nucleosomes containing methylated DNA; B: During DNA semi-reserved replication, DNMT1 interact with the DNA polymerase processing factor proliferating cell nuclear antigen (PCNA) and ubiquitin-like plant homeodomain and RiNG finger domain containing protein 1 (UHRF1) and methylate the hemimethylated sites; C: Soon after DNA semi-reserved replication, DNMT3 correct methylation sites missed by DNMT1 and complete the process.
- Citation: Gao S, Wang K. DNA methylation in liver diseases. World J Clin Infect Dis 2014; 4(4): 41-49
- URL: https://www.wjgnet.com/2220-3176/full/v4/i4/41.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v4.i4.41