Copyright
©2011 Baishideng Publishing Group Co.
World J Clin Oncol. Jan 10, 2011; 2(1): 1-7
Published online Jan 10, 2011. doi: 10.5306/wjco.v2.i1.1
Published online Jan 10, 2011. doi: 10.5306/wjco.v2.i1.1
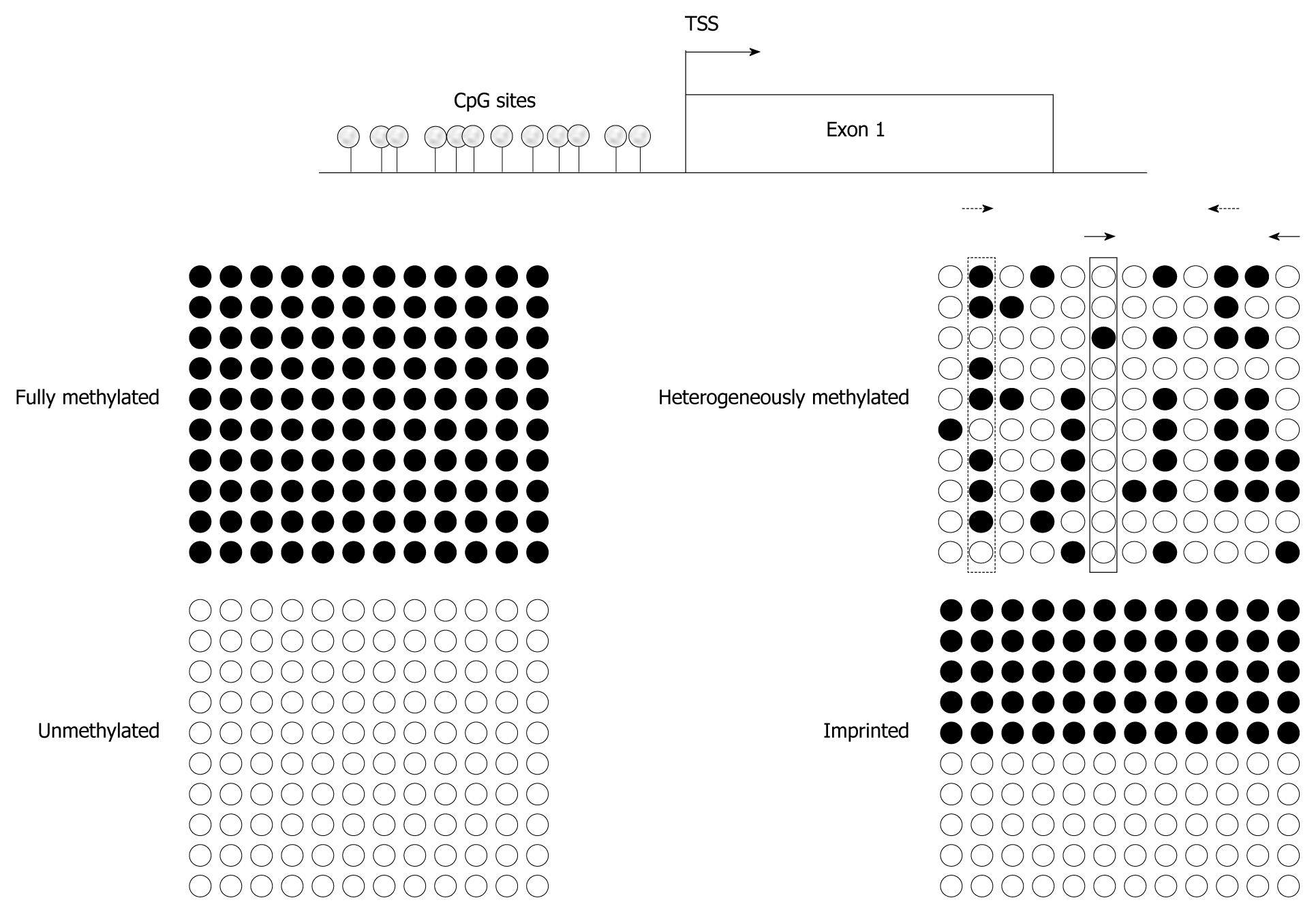
Figure 1 Assaying various methylation states by different techniques.
Idealized bisulfite sequencing results of cytosine-guanosine (CpG)-sites in a proximal promoter region (TSS denoting the transcriptional start site), showing fully methylated, unmethylated, heterogeneously methylated and typical imprinted (differentially methylated) states. Each circle corresponds to a single CpG site, each line to one cloned allele. Filled circles represent methylated and empty circles unmethylated cytosines. Bisulfite sequencing gives an excellent indication of methylation state at every single CpG site, but quantification requires an adequate number of investigated clones. Pyrosequencing, methylcytosine-binding domain (MBD)- or methylation-specific polymerase chain reaction (MS-PCR)-based assays will detect 100% methylation at fully methylated and 0% methylation at unmethylated sequences, respectively. Imprinting results in methylation values of 50% at every single CpG site in pyrosequencing. In addition, MS-PCR-assays will detect signals for methylated and unmethylated DNA. For heterogeneously methylated DNA, bisulfite sequencing yields quite clear results, if a sufficient number of clones are used. In contrast, pyrosequencing will identify high methylation values (dotted rectangle) for several CpG sites and low values (solid rectangle) for other sites, whereas MBD-based analyses lead to intermediate overall methylation values. The results of MS-PCR methods depend critically on the chosen CpG-sites and may vary enormously even with small changes in the PCR stringency. For example, MS-PCR assay using primers interrogating CpG sites 2 and 10 (dotted arrow) yields higher methylation values compared to an assay aimed at CpG sites 6 and 12 (solid arrows).
- Citation: Schulz WA, Goering W. Eagles report: Developing cancer biomarkers from genome-wide DNA methylation analyses. World J Clin Oncol 2011; 2(1): 1-7
- URL: https://www.wjgnet.com/2218-4333/full/v2/i1/1.htm
- DOI: https://dx.doi.org/10.5306/wjco.v2.i1.1