Copyright
©The Author(s) 2020.
World J Clin Oncol. Nov 24, 2020; 11(11): 918-934
Published online Nov 24, 2020. doi: 10.5306/wjco.v11.i11.918
Published online Nov 24, 2020. doi: 10.5306/wjco.v11.i11.918
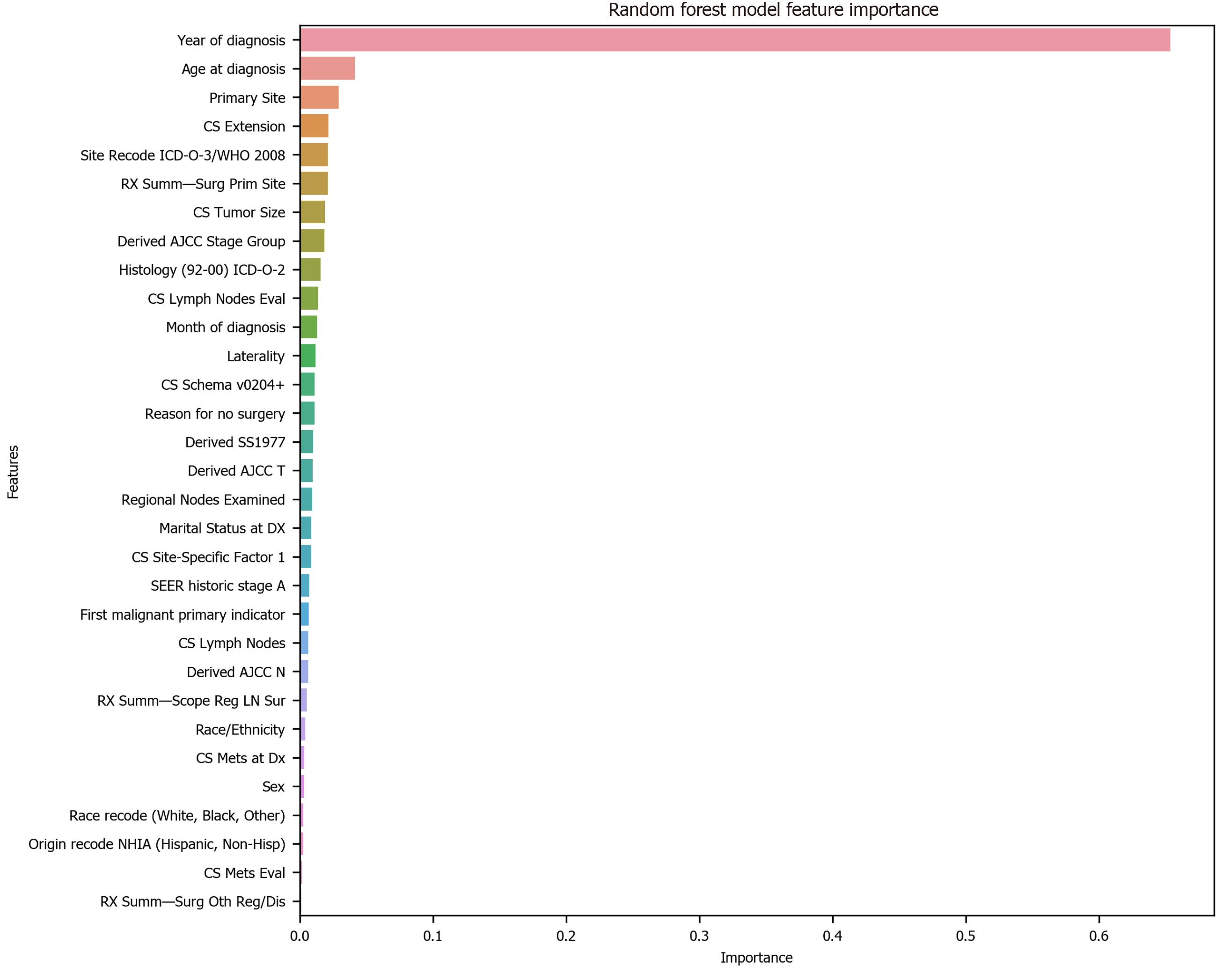
Figure 1 Feature selection using random forest.
CS: Coding system; ICD-O-3: International Classification of Diseases for Oncology, 3rd ed; WHO: World Health Organization; AJCC: American Joint Committee on Cancer; SEER: Surveillance, Epidemiology, and End Results; LN: Lymph node.
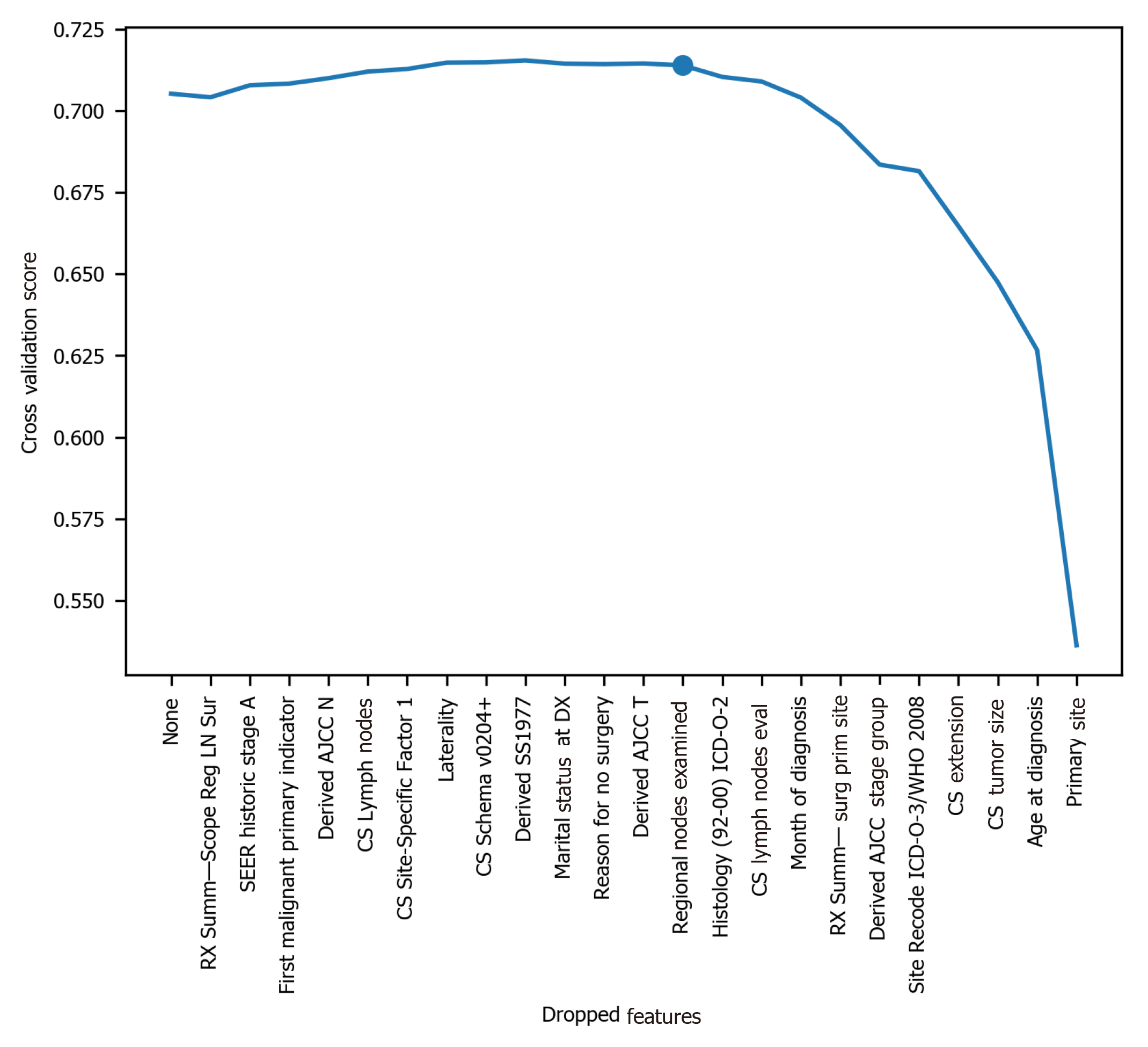
Figure 2 Cross-validation score change for selecting optimal number of features.
LN: Lymph node; SEER: Surveillance, Epidemiology, and End Results; AJCC: American Joint Committee on Cancer; CS: Coding system; ICD-O-3: International Classification of Diseases for Oncology, 3rd ed; WHO: World Health Organization.
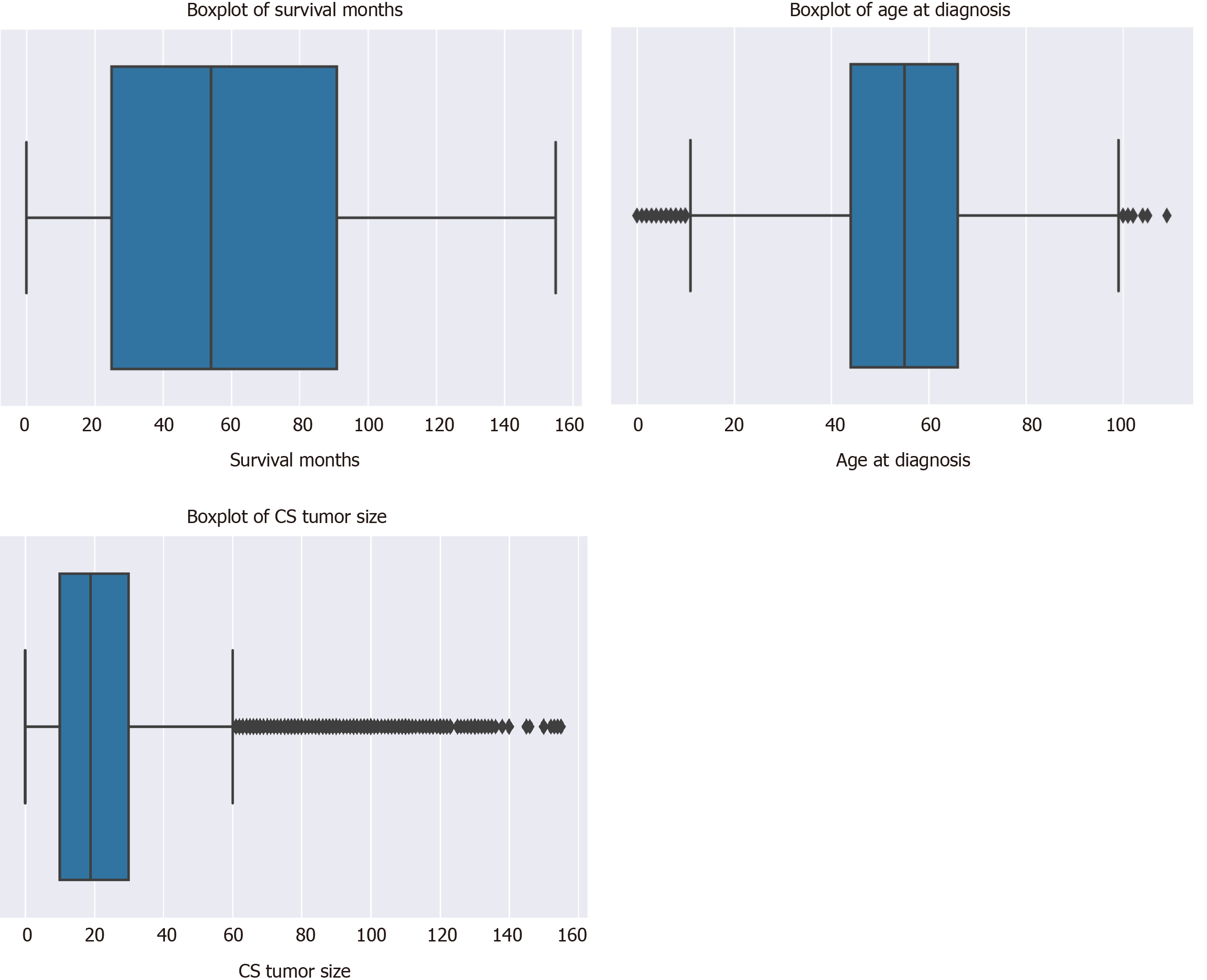
Figure 3 Boxplots of sample characteristics.
CS: Coding system.
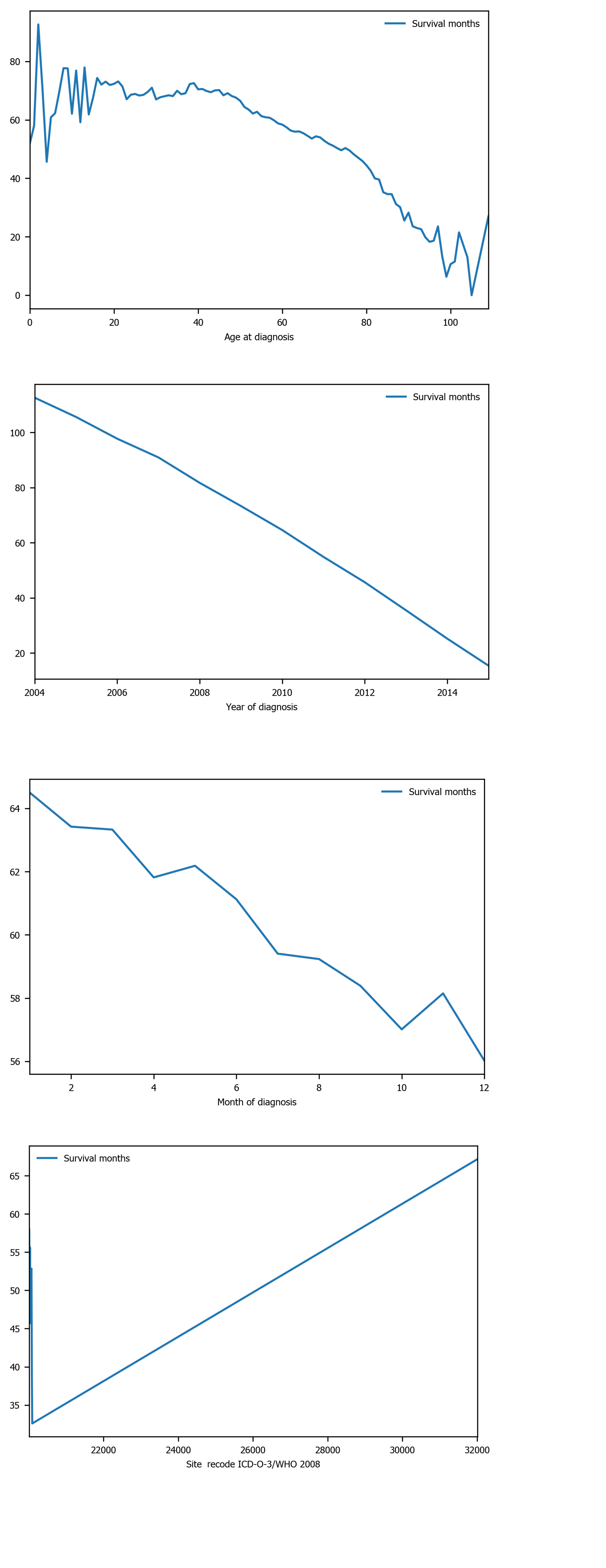
Figure 4 Survival months shows strong linear relation with several variables: Age of diagnosis, year of diagnosis, month of diagnosis, and site recode ICD-O-3/WHO 2008.
ICD-O-3: International Classification of Diseases for Oncology, 3rd ed; WHO: World Health Organization.
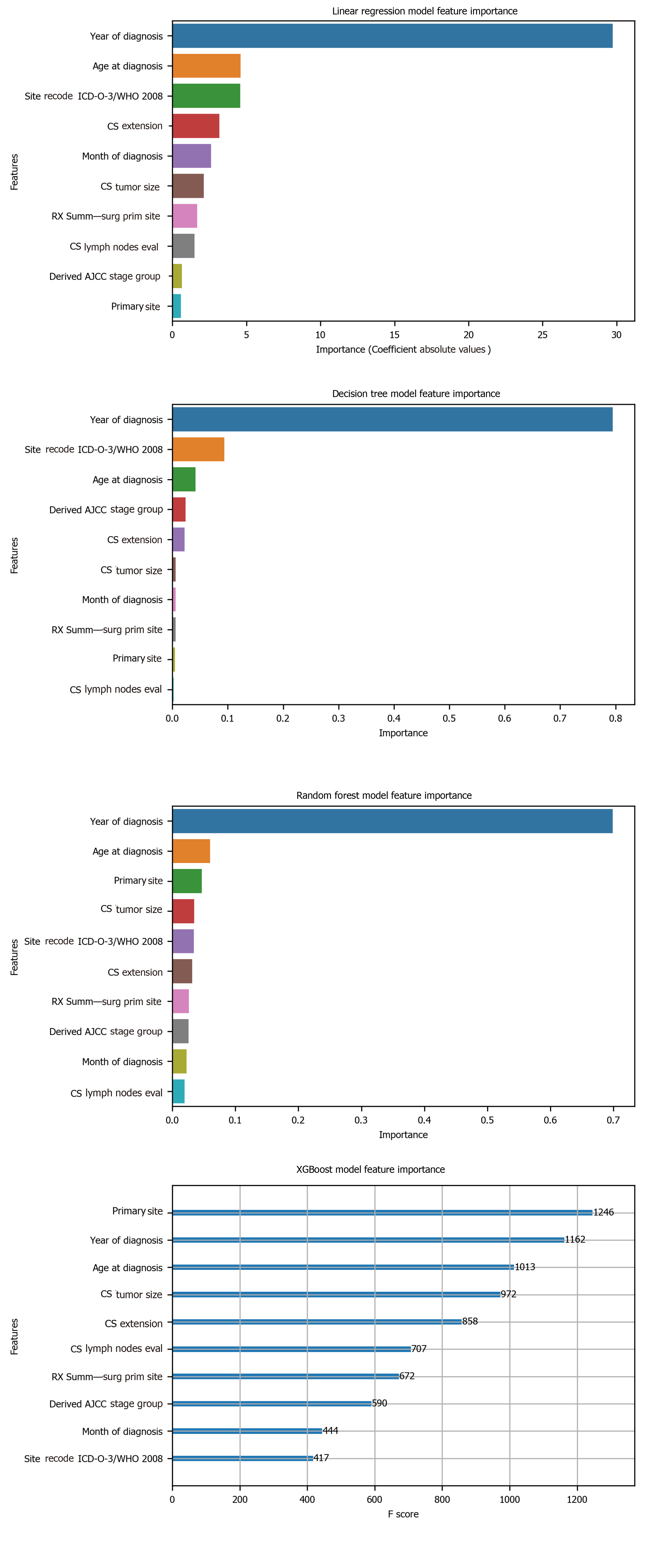
Figure 5 Machine learning model feature importance.
ICD-O-3: International Classification of Diseases for Oncology, 3rd ed; WHO: World Health Organization; CS: Coding system; AJCC: American Joint Committee on Cancer.
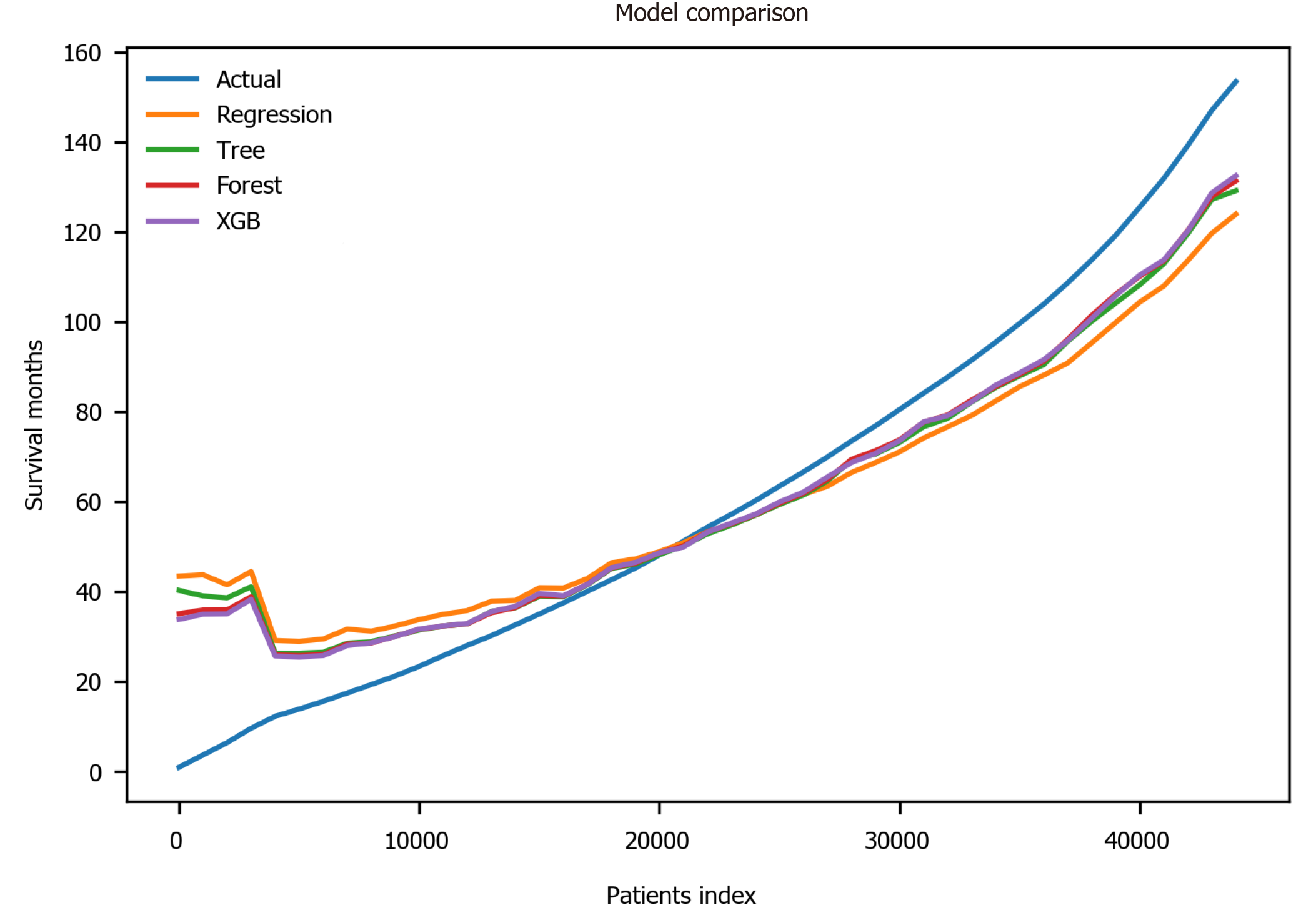
Figure 6 Prediction comparison among different models.
Patient index refers to the rank after sorting by survival months. Actual: The actual survival outcome; XGB: Extreme gradient boosting.
- Citation: Hung M, Park J, Hon ES, Bounsanga J, Moazzami S, Ruiz-Negrón B, Wang D. Artificial intelligence in dentistry: Harnessing big data to predict oral cancer survival. World J Clin Oncol 2020; 11(11): 918-934
- URL: https://www.wjgnet.com/2218-4333/full/v11/i11/918.htm
- DOI: https://dx.doi.org/10.5306/wjco.v11.i11.918