Published online Nov 15, 2021. doi: 10.4251/wjgo.v13.i11.1561
Peer-review started: February 22, 2021
First decision: June 16, 2021
Revised: June 29, 2021
Accepted: September 7, 2021
Article in press: September 7, 2021
Published online: November 15, 2021
In the world, among all type of cancers, colorectal cancer (CRC) is the third most commonly diagnosed in males and the second in females. In most of cases, (RP1) patients’ prognosis limitation with malignant tumors can be attributed to delayed diagnosis of the disease. Identification of patients with early-stage disease leads to more effective therapeutic interventions. Therefore, new screening methods and further innovative treatment approaches are mandatory as they may lead to an increase in progression-free and overall survival rates. For the last decade, the interest in extracellular vesicles (EVs) research has exponentially increased as EVs generation appears to be a universal feature of every cell that is strongly involved in many mechanisms of cell-cell communication either in physiological or pathological situations. EVs can cargo biomolecules, such as lipids, proteins, nucleic acids and generate transmission signal through the intercellular transfer of their content. By this mechanism, tumor cells can recruit and modify the adjacent and systemic microenvironment to support further invasion and dissemination. This review intends to cover the most recent literature on the role of EVs production in colorectal normal and cancer tissues. Specific attention is paid to the use of EVs for early CRC diagnosis, follow-up, and prognosis as EVs have come into the spotlight of research as a high potential source of ‘liquid biopsies’. The use of EVs as new targets or nanovectors as drug delivery systems for CRC therapy is also summarized.
Core Tip: New efficient screening and treatment approaches are strongly mandatory to increase colorectal cancer (CRC) patients’ prognosis. Extracellular vesicles (EVs) represent a promising mean to diagnose and treat colorectal cancers. This review summarizes the most recent literature on the use of EVs in the management of CRC.
- Citation: Mammes A, Pasquier J, Mammes O, Conti M, Douard R, Loric S. Extracellular vesicles: General features and usefulness in diagnosis and therapeutic management of colorectal cancer. World J Gastrointest Oncol 2021; 13(11): 1561-1598
- URL: https://www.wjgnet.com/1948-5204/full/v13/i11/1561.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i11.1561
In the world, colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females. In 2018, 1.8 million new cases were reported with almost 861000 related deaths according to World Health Organization[1]. In Europe and United States, approximately 748000 and 148000 new cases of large bowel cancer are diagnosed annually, two third being colon cancers, the remainder being rectal ones[2,3]. Respectively 242000 and 53000 died of CRC-related diseases. While still treated first by surgery and chemotherapy, despite a better understanding of its natural history and the development of new therapies (immune checkpoint inhibitors, etc.), CRC recurrence and metastasis are still the main causes of death[4]. Thus, determining relevant factors involved in disease progression is strongly mandatory to drive development of new effective strategies for therapies against CRC, etc.). In tumor evolution, recent studies have shown the weight of continuous interplay between surrounding cells (cancer cells with themselves, cancer cells with stromal cells[5]. Such communication strategies require specific mechanisms including direct cell to cell contacts but also autocrine, juxtacrine, paracrine and even endocrine secretion of specific factors (growth factors, matrixins, cytokines, chemokines, etc.)[6]. Among such secreted means figure extracellular vesicles (EVs), a generic consensus term used to describe any type of lipid bilayer-delimited particles, unable to replicate, and extracellularly released by every cell (including microorganisms)[7-9]. EVs surface receptors allow their targeting and capture by a broad range of recipient cells that will incorporate either proteic, lipidic, or genetic messages resulting in modifications of their physiological behavior. These EVs have been recently proved to be efficient communication means in human diseases[10], especially in cancer. As the field of EVs is extremely active[11,12], we aimed to review the respective roles of colonic cells EVs as well as stromal derived-EVs in colon cancer to better understand cellular and molecular mechanisms underlying its occurrence and development. We also underline EVs as powerful and early tools to diagnose colon cancer, to accurately define its aggressiveness, and to better design, in a personalized approach, treatment strategies.
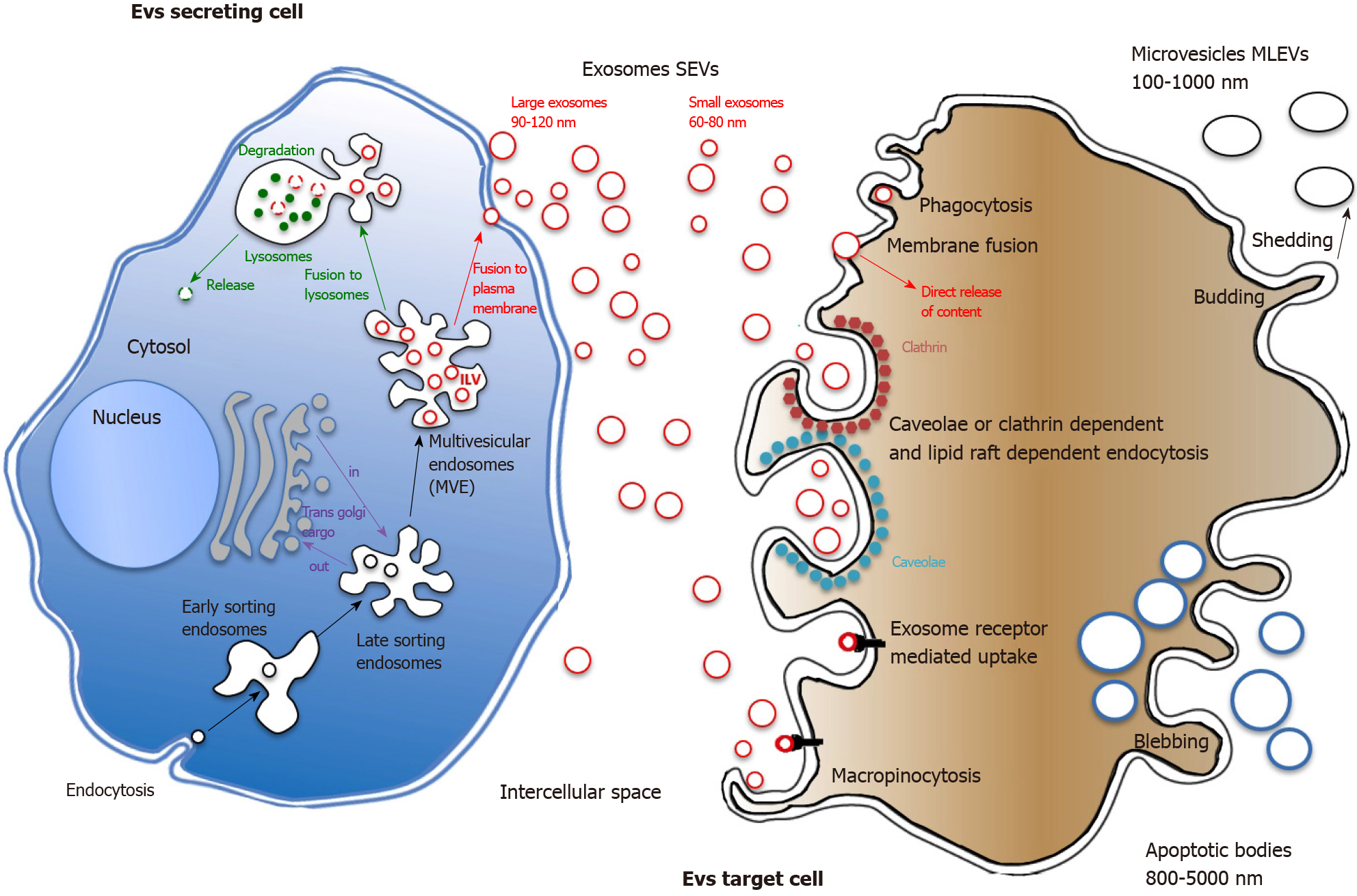
Either eukaryotic or prokaryotic cells produce continually various amounts of 40-1000 nm membrane vesicles that are released into local environment. Such EVs can be evidenced in the conditioned media of every cultured cell, but also in almost all biological fluids (including blood, cerebrospinal fluid (CSF), urine, saliva, seminal plasma, and breast milk)[13,14]. EVs definition embodies different terms, sometimes used indifferently in literature, including exosomes, microvesicles, microparticles, multivesicular bodies, apoptotic particles, apoptotic bodies, oncosomes, etc. As not yet defined biomarkers can specifically categorize each vesicle, as a rule the 2018 minimal information for studies of extracellular vesicles consensus recommends to label bilayered vesicles smaller than 200 nm as small EVs (SEVs) and those larger than 200 nm as medium large EVs (MLEVs)[15]. Alternatively, the original process of the cell can also be mentioned: Oncosomes specifically refer to oncogene containing EVs, large oncosomes being massive EVs (over 1000 nm) produced by oncogenically transformed cells[16]. As they lack bilayered membrane, this definition should exclude the recently discovered sub-50 nm nanoparticles exomeres[17].
MLEVs production: MLEVs, so called ectosomes, are heterogeneous membranous vesicles generally originating from outward plasma membrane budding (ectosomal release)[18]. In contrast with apoptotic bodies or necrotic blebs of the plasma membrane (PM) that are the consequences of complex structural transformations resulting in dying cells disassembly[19], ectosomes are shed by living cells.
SEVs synthesis & release: Unlike ectosomes, SEVs stemmed from the endosomal compartment. SEVs biogenesis starts with the inward budding of small portions of the plasma membrane containing outer membrane exposed material. These small intracellular vesicles form the early endosome. Inward budding of the limiting membrane of the early endosome then occurs, resulting in the progressive assemblage of intraluminal bilayered vesicles (ILVs) within so-called large multivesicular endosomes (MVEs) (Figure 1). During this process, cytosolic proteins as well as nucleic acids can be trapped into ILVs through the action of the endosomal sorting complex required for transport (ESCRT) machinery[20]. ESCRT is a family of proteins that associate in successive complexes (ESCRT-0, -I, -II and -III) at MVEs membrane to sort ubiquitinated cargos into late endosomes[21]. ESCRT is also essential for ILVs generation and cargo targeting driving through deubiquitinating enzymes recruitment[22,23]. Interestingly, such protein sorting can also follow a ceramide ESCRT-independent pathway suggesting a critical role for lipid raft microdomains in MVEs formation[24]. Most of MVEs are further directed for cargo degradation into lysosomes by fusing with them. Nevertheless, MVEs also contain intralumenal proteins and lipids, which are not intended for lysosome degradation. ILVs can release their content into the cytoplasm by undergoing direct back-fusion with the endosome limiting membrane[25]. Progressive acidification along the endocytic pathway seems to be required for degradation and recycling of internalized components suggesting that pH could be a major determinant of MVEs degradation vs secretion functions[26]. Indeed, concerning MVEs secretory function, a subset of MVEs fuse to PM and release their content into the extracellular space, in the form of SEVs, a process called exosome biogenesis[27]. MVEs that are fated for exocytosis are transported to PM along microtubules by the molecular motor kinesin[28]. MVEs docking to PM are strongly regulated by the Rab family of small GTPases proteins. Depleting Rab27a prevented MVEs to efficiently fuse with the PM while Rab27b knockdown resulted in perinuclear MVEs accumulation, both observations suggesting that Rab27 was responsible for trafficking MVEs to the cell surface[29]. Once docked, secretory MVEs couple to the SNARE (soluble N-ethylmaleimide-sensitive component attachment protein receptor) membrane fusion machinery[30]. SNARE complex formation and membrane fusion are tightly controlled by multiple regulatory mechanisms[31] among which figure phosphorylation profile of SNARE proteins that influence either SNARE complex localization or interaction with SNARE partners[32].
EVs capture: Once released by the secreting cell, EVs distribute to extracellular matrix (ECM) then circulate locoregionally or distantly to deliver their molecular cargo to recipient cell. EVs cargo is protected from degradation and is rapidly taken up by different organs, such as liver, spleen and lymph nodes[33]. Circulating labelled EVs half-life has been evaluated in mice to be about 2 min but it remains possible to detect EVS in the bloodstream hours after injection[34]. Although still globally unknown, differences in EV size and presence of outer surface membrane components probably could account for their recognition and capture by target cells[35]. Once recognized, strongly depending on recipient cell type[36], EVs will enter through a variety of endocytic routes, either through clathrin dependent or independent pathways (caveolin-mediated uptake, lipid raft-mediated internalization, etc.). Also, both phagocytosis and macropinocytosis can been involved in EVs uptake[37], the latter being very efficient for specific EVs like those harboring CD47 at their surface[38]. After internalization, while endosome seems one of the best candidate locations for EVs membrane fusion then cargo delivery, EVs intracellular fate remains a matter of debate (Figure 1).
Altogether, due to the multiple sorting mechanisms that determine specific molecules incorporation into EVs, the distinct vesicle subpopulations carrying different cargo that can be evidenced, and the complex pathways/factors that regulate EVs export and secretion, EVs biogenesis threshold is likely to greatly vary between cell types according to their physiological/pathological status. The high rate of SEVs secretion found in transformed cells suggests that the balance between EV degradation and secretion is disrupted in cancer towards EVs cargo release[39]. This kind of change is not specific to cancer cells but may also occur in non-transformed cells. In antigen-presenting cells, large amounts of SEVs are found to be released upon stimulation[40].
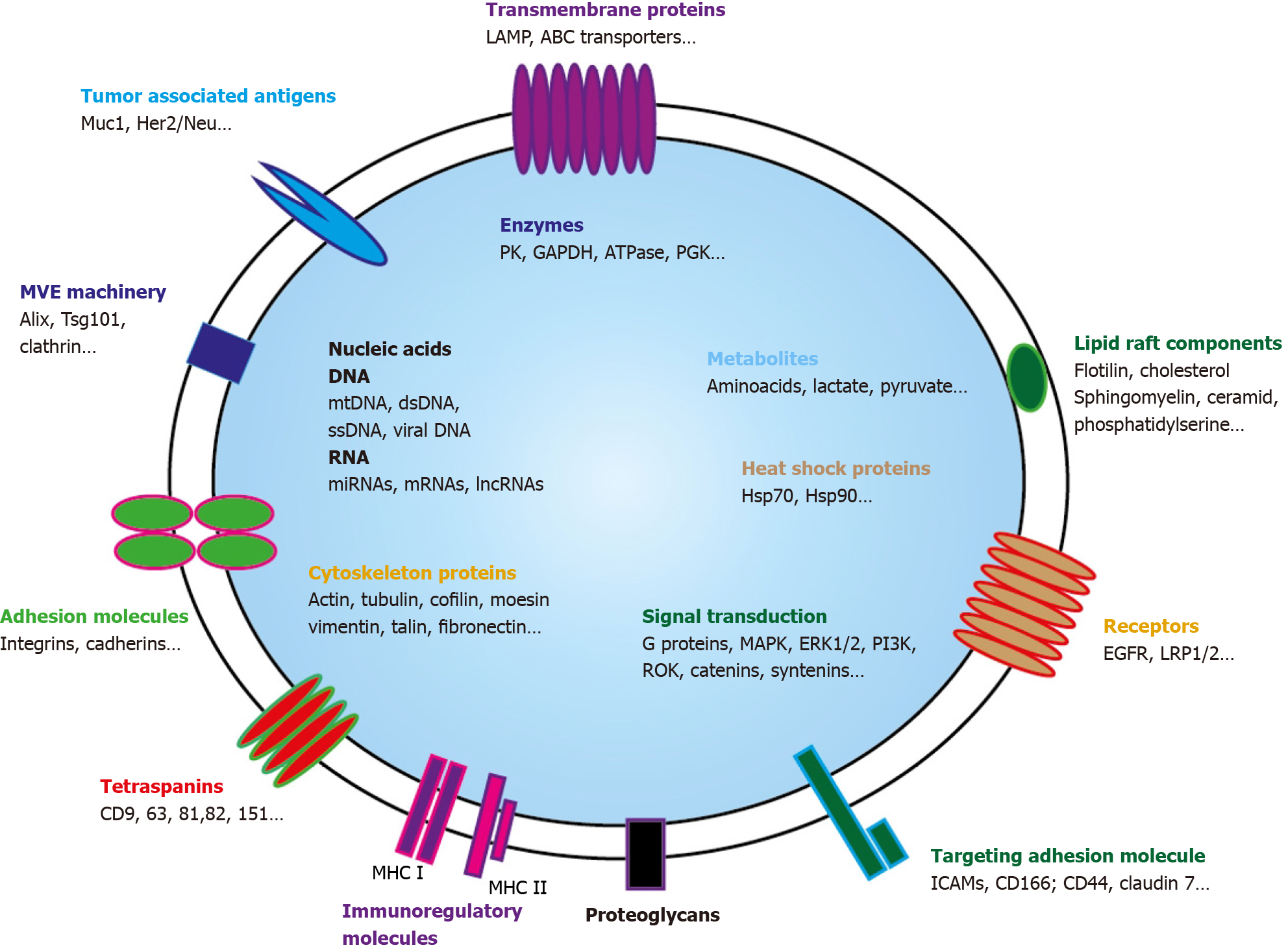
EVs are highly heterogeneous and likely reflect the phenotypic state of the cell that generates them[41]. Every EVs behave as a multi-molecular cargo whose bilayered membranes regulate its stability by protecting bioactive content from degradation[42]. Alike cells, EVS can contain inside their lipid bilayer every basic constituent of a cell including metabolites[43], functional proteins (enzymes, receptors, transporters, etc.)[44-46], but also nucleic acids molecules such as mRNAs[47], interfering microRNAs (miRNAs)[48], small and long non-coding RNAs (snRNAs & lncRNAs)[49], and even mitochondrial DNA[50] or more recently genomic DNA[51] (Figure 2).
EVs protein cargo: Because of their endosomal origin, and since they derived from the ILVs in MVEs, SEVs biogenesis is heavily dependent on the mechanisms that regulate MVEs maturation and trafficking. SEVs mostly contain proteins originating from the cytosol and either endosomes then PM components[7]. As budding and release of EVs require inner PM actin polymerization then actomyosin cytoskeleton contraction, cytoskeleton proteins such as actin and tubulin are generally found in EVs[52,53]. Among highly representative proteins that can also be found in SEVs figure important regulators of EVs trafficking: (1) Members of the Rab family that play well-established roles in vesicle transfer between intracellular compartments such as MVEs driving to PM for SEVs secretion[54,55]; (2) SNARE membrane fusion machinery, through SNARE complexes recruitment, that is specifically required for MVEs docking then fusion with PM[30,35,56]; (3) ESCRT proteins and important ESCRT side molecules implicated in ESCRT assembly or nucleation like ALIX[57]; and (4) Tetraspan transmembrane proteins (tetraspanins), highly enriched in SEVs, that are also involved in ESCRT-independent EVs release[58,59]. Tetraspanins display high affinity for cholesterol and sphingolipids such as ceramides which may create PM microdomains as it occurs in membrane reconstitution experiments[60]. Their interaction with PM proteins, either by direct association or by entrapment in tetraspanin-enriched PM microdomains, facilitates their sorting into EVs[58,61-63].
Interestingly, EVs can also transport mitochondrial proteins that may be active. Two mitochondrial inner membrane proteins MT-CO2 (encoded by the mitochondrial genome) and COX6c (encoded by the nuclear genome) were highly prevalent in the plasma of melanoma patients, as well as in ovarian and breast cancer patients defining a new EVs subtype[64]. As not only mitochondrial membrane proteins but also mitochondrial enzymes are present in EVs, mt-EVs could affect the metabolic output of the recipient cells by either preventing inflammation[65] or promoting tumor growth[66-68].
SEVs specific endosomal-driven content allows their distinction from ectosomes that can directly bud and shed from PM at lipid-raft-like domains[69]. These vesicles, now generically referred to as MLEVs, are extremely heterogeneous in size, ranging from 200 nm to as large as 10 μm. They are generally enriched in cell surface or integral transmembrane proteins, reflecting their PM origin[70,71]. For example, during reticulocyte maturation, autophagosomal exocytic event is coupled with plasma membrane blebbing that release glycophorin A, an integral plasma membrane protein, into budding vesicles[72].
Last, SEVs content is also distinct from apoptotic microparticles or apoptotic bodies (apoBD). ApoBDs are larger than SEVs and MLEVs as they have a diameter of 800–5000 nm[73]. ApoBDs encapsulate residual ingredients of dying cells. They are enriched with autoantigens and pro-inflammatory factors[74,75] and bear key markers of cell disassembly such as ROCK1 and PANX1 and apoptotic markers such as CD31 or Annexin V.
EVs metabolite cargo: Aside proteomic studies that try to unravel the complex protein repertoire in EVs, metabolomic studies reveal that EVs contain different classes of low-molecular-weight compounds. Organic acids, nucleotides, sugars and their derivatives, carnitines, vitamins and related metabolites, and amines are frequently evidenced in EVs[43]. Of course, most of these metabolites were generally derived from cytosolic cellular pathways, as large portions of cytosol are engulfed in ILVs then EVs[76]. Nevertheless, metabolites presence could also result from either specific metabolite sorting or ILVs/EVs in situ synthesis through residing metabolic enzymes as high metabolite concentrations over the cellular levels were reported in EVs[77]. Complete but more often partial metabolic routes can be evidenced in EVs explaining why EVs metabolite identification does not generally cover the whole parental cell metabolome but represents a miniature subset of it.
Lipids are also frequently found in EVs. EVs lipidome analysis allows characterization of different classes of lipids, including glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, and fatty acids confirming similarity between EVs lipid content and their parental cells membranes composition[78]. As it is important to preserve functional flexible lipid bilayer as well as right ion composition and pH-homeostasis[60], numerous ATP-driven transporters and ion-pumps are also found in EVs. To be fully functional, these elements need energy supply that may be given either by glycolytic enzymes[79] or even mitochondrial ATP synthase that is frequently found in EVs[64]. To optimize energy thresholds, such enzymes and substrates seems to be organized in metabolons that have been found to be fully functional in EVs[80].
Every cell may send out a range of messages to distinct still unknown targets, and both messages and targets may vary depending on the metabolic state of the producing cell. In EVs metabolic composition is of importance as it may represent a specific environment (“climate”) the parental cell is going to transfer to the recipient one. By providing substrates for biosynthesis, EVs-transported aminoacids (glutamine, leucine…) have been shown to strongly affect the tricarboxylic acid (TCA) cycle of the recipient cancer cells thus improving nutrient status of fast growing and proliferating cells[81]. By providing both enzymes and substrates, adipocytes EVs stimulate melanoma fatty acid oxidation (FAO) that increase mitochondrial activity redistributes mitochondria to membrane protrusions of migrating cells, which is necessary to increase cell migration[82]. Interestingly, using various cell culture protocols, several reports have shown that EVs production in quantity and composition is largely influenced by external factors[83], the most striking variation being in the EVs metabolomes[84]. As slight metabolic variations could drive cancer cell repro
EVs RNA cargo: Valadi and Skog both demonstrated that EVs transported mRNAs that can be translated into protein, providing the first evidence of virus-independent genetic material horizontal transfer between cells[86]. Since these pioneering studies, the presence of RNAs, within EVs have been reliably shown with either microarrays or real-time quantitative polymerase chain reaction techniques in numerous reports[47]. This presence can easily be explained as cytosolic proteins engulfment, resulting from a microautophagy process[87], involve proteins located close to the MVE outer membrane during its inward budding and can comprise RNAs molecules[86]. Those RNA species include not only mRNAs but also rRNA, tRNA, snRNA, snoRNA, piRNA, Y-RNA, scRNA, SRP-RNA, 7SK-RNA and lncRNAs. All these RNAs can be transferred to the recipient cells[88,89]. In addition, two major components of the RNA-Induced Silencing Complex, namely DICER and Argonaute, aimed at producing miRNAs have been shown to associate with MVE and to be sorted into exosomes[48,90]. This suggests that miRNAs are likely to be packaged into EVs along with proteins required for their processing or function)[91]. As largely protected from RNAses when packaged in EVs, miRNAs driven-gene regulation will be able to generate a multifaceted signaling response in the target cell. As EVs mRNAs are also functional and can be translated in the target cell[86], both mechanisms provide a direct modulation of recipient cell protein production. This new signaling pathway play specific roles in intercellular communication during various physiological[14,92] or pathological processes. Indeed, numerous reports have described the ability of EVs RNAs to impact the functional properties of cells that incorporate them[93], especially in the cancer field where such mechanism may drive apoptosis resistance[94], drug resistance[67,95,96], and metastatic behavior[89].
EVs DNA cargo: Extracellular DNA is present in the circulation and may represent an attractive marker issue for liquid biopsies. In plasma, DNA is found both in free form and enclosed in EVs[97,98]. Rather than being packaged within EVs membrane-bound space, DNA seems mostly attached to the outer surface of EVs[99]. Quantities as well as properties of packaged DNA may largely vary in different subsets of EVs even originating from the same source. It is likely that the heterogeneity of DNAs in EVs is related to the size of EVs. In contrast to SEVs that are more frequently devoid of DNA, large size intact DNA (> 2 Mbp), generally associated to histones, is commonly found in LEVs[100,101]. EVs DNA fragments may represent and even cover all chromosomes of parental cells[51,97]. As DNA sometimes harbor mutations, it may reflect the mutational status of parental DNA[102-104] and thus serve as a relevant oncologic biological marker.
Beside single stranded and/or double stranded genomic DNA, mtDNA can also be found in EVs extracted from cell culture medium[105,106] but also in plasma EVs[107] where presence of complete mitochondrial genome has been evidenced. Transfer of this complete mtDNA molecule seems to drive recipient cells fate[108].
Considering the many cell types that interact at the mucosal interface, the intestinal lumen could be a rich source for EVs in large bowel tissues as well as an interesting source of disease-specific EVs in pathological conditions.
Normal colonic cells as a primary source of EVs: As most of our tissues, colonic tissue may be an important source of EVs. Intestinal epithelial cells (IEC) are located at the strategic interface between external environment and the body most extensive lymphoid compartment. Aside their essential role in nutrients absorption, IEC have been shown to play a key role in immune response by promoting and regulating luminal antigens presentation to mucosal immune cells[109] through EVs release at both apical and basolateral sides as IEC display all the elements needed for either antigen processing or EVs production[110]. These EVs contain molecules that are implicated in adhesion and antigen presentation, such as major histocompatibility complex (MHC) class I molecules, MHC class II molecules, CD63…[111]. As these EVs may also contain CD133, whose presence in lipid rafts play a pivotal role in the maintenance of stem cell features[112], it has been suggested that CD133-containing EVs release may contribute to cell differentiation by reducing and/or modifying stem cell characteristic membrane microdomains composition within IEC apical plasma membrane[113].
Maintenance of the intestinal stem cell can be driven by niche-derived EVs: The intestinal epithelium is continuously renewed by a small proliferating intestinal stem cell (ISC) population residing at the bottom of the intestinal crypts in a specific microenvironment, the stem cell niche[114]. Niche surrounding cells including intestinal subepithelial myofibroblasts, endothelial cells and macrophages, generate Wnt, Notch, hedgehog and epidermal growth factor (EGF) signals that maintain ISC as a stem cell[115,116]. Mutations within these key signaling pathways can deregulate ISCs from the control of regulatory signals, allowing them to develop precursor lesions[117]. Once induced, intestinal regeneration through ISC symmetric division is strongly dependent on specific signals such as the recently evidenced IL-22[118]. In that intestinal homeostasis general regulatory process, EVs can also largely participate as intestinal fibroblast-derived EVs are involved in forming the ISC niche by transmitting Wnt and EGF activity[119] as well as intestinal macrophage-derived EV-packaged Wnt are essential for regenerative response of intestine against radiation[120]. EVs can also drive ISC differentiation as Rab8a vesicles regulate Wnt ligand delivery then Paneth cell maturation at ISC niche[121]. Such EVs-driven mechanism has also been shown to impose quiescence on residual hematopoietic stem cells in the leukemic niche[122].
Microbiota as an important source of EVs: Intestinal tract is a specific place where communication between many different species (bacteria, fungi, parasites…) occurs continually. Not only human IEC but also commensal bacteria are known to release signaling vesicles[123]. Interestingly, many studies have shown that intestinal microbiota can be shaped either by food plant-derived EVs[124] or host-derived EVs[125] suggesting multidirectional influences on each other of all intestinal tract living species. Such interspecies communication has also been evidenced between resident helminths and host IEC[126,127]. Every bacteria, parasite, fungi… generate a huge reservoir of antigen that can induce host immune response. Thus, once initiated, this response can be tailored through complex cross reacting EVs modulation leading to either immune tolerance or inflammatory reaction.
Numerous studies have demonstrated that circulating EVs increased in patients with intestinal pathologies while EVs fractions are different in cancers, compared to patients with inflammatory intestinal diseases such as Crohn's or inflammatory bowel diseases (CD or IBD)[128].
EVs deregulation in intestinal inflammatory diseases: Chronic inflammation pathologies of gastrointestinal (GI) such as IBD, CD, Helicobacter pylori-associated inflammation and chronic pancreatitis have been identified as strong risk factors for cancer development[129]. Interaction of different genetic, microbiome, and environmental factors with the immune system drives IBD complex characters. The balance between immune suppression and stimulation against environmental factors is largely disturbed in IBD patients, resulting in inflammation and compromised integrity of the intestinal barrier. Elevated levels of EVs and/or EV content have been identified in IBD patients. EVs can modulate the immune response[130]. Among immune cells, macrophages are essential for the maintenance of intestinal homeostasis[131]. Serum EVs isolated from the dextran sulphate sodium-induced acute colitis mouse model could activate macrophages[132]. as well as EVs derived from the colonic luminal fluid of IBD patients that contained high mRNA and protein levels of several inflammatory cytokines could promote macrophage migration[133]. Dysfunction of regulatory T cells (Tregs) has been shown to be associated with a failure of intestinal tolerance, and contributes to the pathogenesis of IBD[134]. EVs derived from Tregs were shown to induce other T cells to develop into the Treg phenotype[135].
EVs release in colorectal cancer: Acidity and hypoxia are key features in cancer that could affect exosome release. Tumor pH may range from 6.0 to 6.8, and the level of acidity is directly associated to the tumor level of malignancy as it selects among cancer cells those that will resist[136]. One consequence of acidity-driven cancer cell selection pressure is an increased EVs release by human cancer cells[137,138].
Hypoxia is also a common characteristic of solid tumors and is associated with cancer progression and poor outcomes. It is generally associated with hypoxic environment that has also been shown to be an important cause of EVs release[139]. Hypoxic CRC cells can transfer Wnt4 mRNA to normal CRC cells by exosome, which can activate β-catenin signal and potentiate the invasive ability of normal CRC cells[140]. In hypoxic microenvironment, CRC cells-secrete miR-410-3p in EVs that promotes progression and metastatic potential of normoxic CRC cells via PTEN/ PI3K/Akt pathway[141].
Epithelial cancers may be driven by a relatively rare sub-population of self-renewing, multipotent cells, named cancer stem cells or cancer-initiating cells (CSCs). Increasing data show that CSCs play a crucial role not only in primary colorectal tumor formation but also in metastasis[142]. In addition, CSCs play a critical role in CRC relapse[143]. They display unique properties of self-renewal, infinite division and multi-directional differentiation potential[144]. Asymmetrical growth and slow-cycling cellular turnover renders them resistant to therapies that target rapidly replicating cells[145]. Not all CSCs in primary lesions are metastatic, allowing distinction between stationary cancer stem cells (SCSCs) and migrating cancer stem cells (MCSCs)[146]. SCSCs exist in colonic epithelial tissues and are active even in benign precursor lesions, contributing to tumor mass proliferation in situ[147]. On the contrary, MCSCs, which have undergone EMT, possess motility characteristics and are able to spread in other tissue to form metastatic tumor mass[148,149].
Untreated colorectal tumors contain a population of quiescent/slow cycling cells resembling CSCs and overexpressing EMT markers such as Zeb2[150]. As for ISC, maintenance of these scarce CSCs generally resides in very specialized niches[151], allowing them to stay dormant for various to long periods of time[152,153]. These niches represent a positive specific microenvironment which is able to maintain stemness and pluripotency[154]. The release of EVs by mesenchymal stromal niche surrounding cells drive hematopoietic stem cell clonogenic potential maintenance and survival, by preventing apoptosis through EV gene expression regulation[155].
This continuous crosstalk between CSC and their surrounding microenvironment is critical as a tiny variation in its modulation could induce important deregulation and subsequent tumor progression[156]. For example, miR-196b-5p, which is highly enriched in CRC patients serum EVs[157] has been shown to promote either CRC cells stemness or chemoresistance to 5-fluorouracil (5-FU) via targeting negative regulators of the STAT3 signaling pathway. Understanding the importance of EVs transfer in that context is a key feature for future CRC therapy[158].
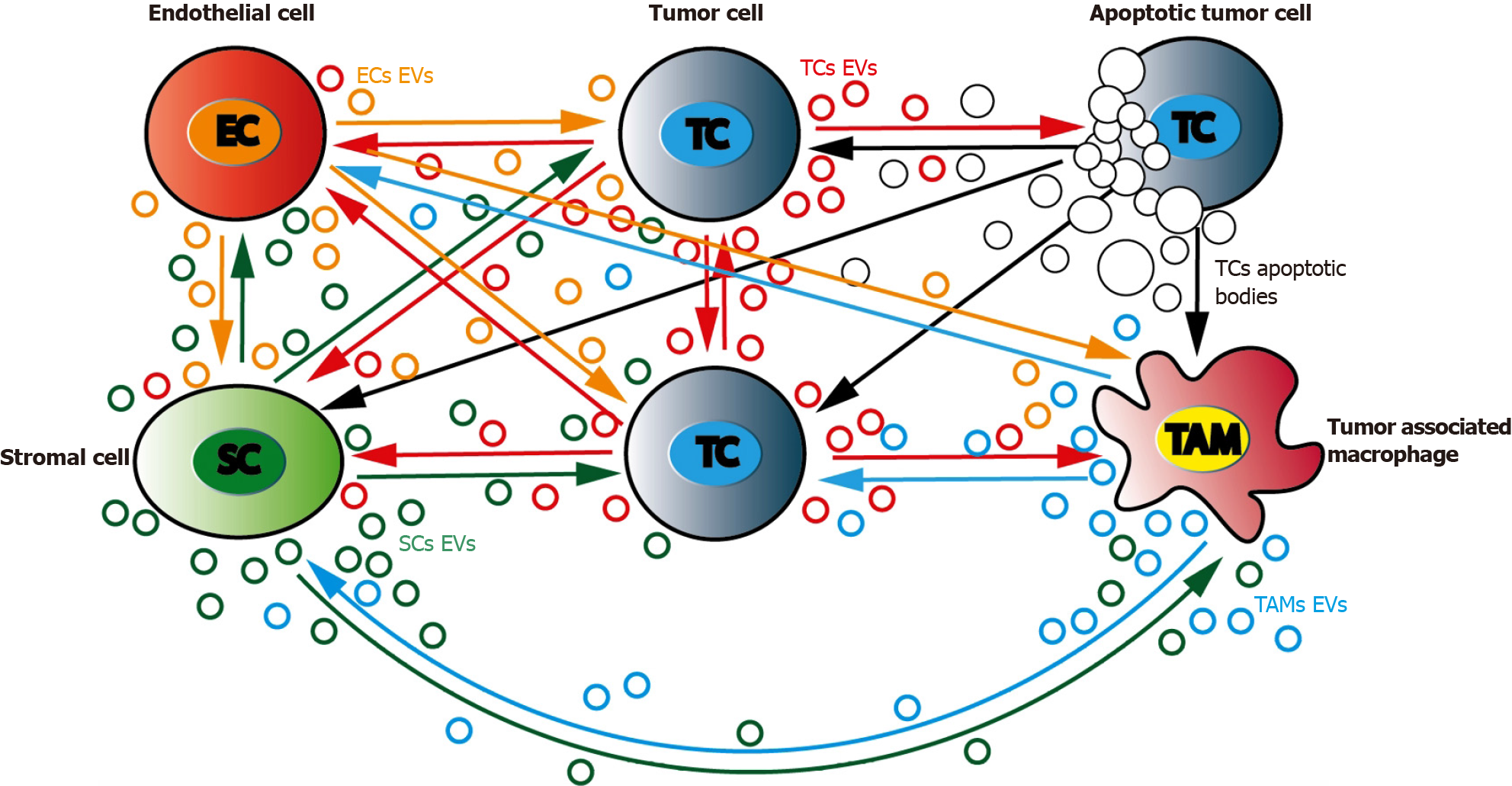
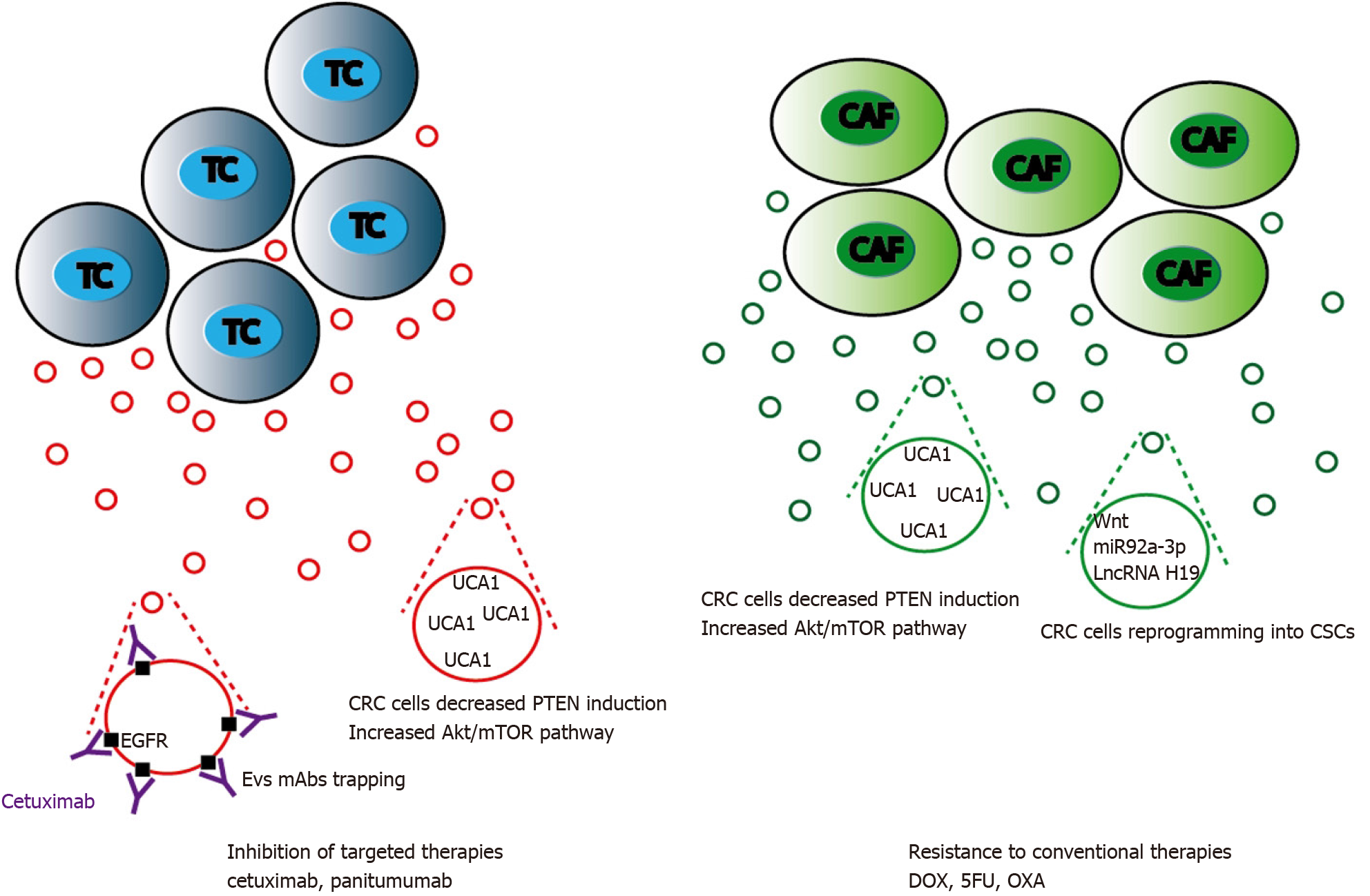
Tumor microenvironment (TME) is a complex and dynamic network including both cancer and stromal cells. Stress conditions such as hypoxia, starvation, and acidosis increase tumor cells EVs release leading to TME changes and expansion. Such specific behavior is the consequence of a complex combinatory of bioactive molecules present in EVs[159]. Not only different form of RNAs but also proteins or lipids could account for these important changes. The release of CD133+ EVs by poorly differentiated CRC cells was found to increase Src and ERK phosphorylation in surrounding cells, with subsequent MAPK intracellular signaling activation and promotion of tumor growth[113]. In response to CRC cells, TME modifications induce EVs-driven stromal cells response that subsequently results in tumor progression by further modifying CRC cells[160]. This continuous dual EVs-driven interplay between stromal and CRC cells is central in tumor behavior as it may drive either tumor cells proliferation or migration[161] (Figure 3).
Among TME, fibroblasts such as cancer associated fibroblasts (CAFs), endothelial cells and infiltrating immune cells are likely to be the major cell types that interacts with tumor cells through EVs signaling[162,163]. Both nature and composition of TME-derived EVs is of importance as cellular origin of the EVs cargo will determine specific changes within the recipient cell[164]. Analyzing their effect on CRC tumor cells, TME-originating EVs have been evidenced to play a central role in cell proliferation[165], acquisition of invasive properties and increased migration[166,167], resistance to chemotherapy[168], angiogenesis development[169], and escape from the immune system.
On the other side, several tumorigenic signals are derived from CRC cells and conveyed to stromal cells through EVs. From the very beginning of CRC progression, CRC cells secrete EVs that can deeply modify TME cells[170]. CAFs are prompted by CRC cells EVs to harbor a highly pro-proliferative and pro-angiogenic phenotype[171]. These important stromal changes are driven by CRC cells EVs composition that is itself largely modulated by different factors such as differentiation or hypoxia[113].
Accumulated genetic and epigenetic changes often activate the expression of oncogenes while silencing tumor suppressors during carcinogenesis. In CRC, several protooncogene mutations affecting KRas, BRaf, PTEN, PIK3CA or TP53 are now well known to promote CRC cells proliferation through cell cycle key players deregulation[172]. Interestingly, mutant KRas expression in donor cell alter EVs cargo composition[173,174]. Such KRas mutation can be transferred through EVs cargo to non-transformed neighboring recipient cells leading to enhanced growth of these newly KRas-expressing cells[175]. However, aside these genetic transfers, most of the profound changes that drive cancer cell proliferation remains of epigenetic origin. Many different mechanisms can be used to alter gene expression, among which figure transfer of EVs cargo content that can increase cell proliferation by their oncosuppressive properties[176]. By suppressing fibroblast TP53 expression, CRC cells EVs miRNAs promote tumor progression[177]. This holds also true for DeltaNp73 enriched EVs that promote oncogenic potential of recipient cells[178]. Such CRC cells EVs transfer can play a role in a synergistic manner with classical factors acting on CRC cell growth in a paracrine manner[179].
All along the natural history of cancer, malignant cells should exhibit high metabolic plasticity to adapt themselves to tumor and surrounding environment continual changes[180]. Tumor cell proliferation continuously demand the highest nutrient capacity to fulfill enhanced biosynthetic and bioenergetics requests. In normal cells, metabolism of glucose is mainly performed through cytosolic glycolysis then mitochondrial TCA and OXPHOS that produce ATP. As mitochondrial PDH is inhibited and pyruvate cannot be transformed into acetyl-coA, cancer cells enhance glycolysis to produce sufficient ATP and generate high lactate content even in aerobic conditions (the “Warburg effect”), both being hallmarks of cancer[181]. High lactate production and release induces TME acidification promoting immune surveillance escape and metastasis[182]. As lipids, amino-acids, and nucleotides are strongly required for cancer cell multiplication, either fatty acids synthesis and FAO[183], or glutamine and serine metabolisms are all increased in tumor cells. Glutamine appears as a major energy substrate in cancer cells. Glutamine could produce TCA cycle intermediates to provide an additional energy source for cancer cells[184]. It has been recently shown that TME metabolism can largely modulate cancer cells progression. CAFs can provide metabolites that will facilitate tumor cells ATP production. Lactate, exported through CAFs MCT4 lactate shuttle then up-taken through cancer cells MCT1 Lactate transporter, could be used to fuel surrounding cancer cells, a process called “reverse Warburg effect”[185-187]. TME can also induce cancer cells FAO through cancer-associated adipocytes free fatty acid (FFA) release then cancer cells FFA CD36 uptake, hereby promoting cancer progression[188]. TME associated endothelial cells that mediated tumor angiogenesis are highly glycolytic[189] while tumor-associated macrophages (TAMs) polarization to immunostimulatory M1 or immunosuppressive M2 phenotype is largely driven by metabolism, M1 cells being highly glycolytic whereas M2 cells mostly relying on FAO and OXPHOS[190]. All these TME cells can shed EVs that will modulate cancer cells metabolism and play a role in their proliferation. EVs can contain metabolites but also metabolism enzymes that can modulate cancer cells metabolism. Uptake of EVs enriched in metabolic enzymes ALDOA and ALDH3A1 accelerated glycolysis thus promoting unirradiated lung cancer cells proliferation[191]. EVs lncRNA SNHG3 sponging miR-330-5p in recipient cells positively regulated pyruvate kinase M expression inhibiting OXPHOS, increasing glycolysis, and promoting breast cancer cells proliferation[192]. As EVs can be produced bi-directionally (Figure 3), cancer cells can also modulate TME cells fate through metabolism reprogramming. Human melanoma-associated EVs miR-210 and miR-155 can reprogram CAFs metabolism to enhance glycolytic phenotype leading to extracellular acidification that favors pre-metastatic niche formation[193]. Prostate cancer cells EVs transfer of PKM2 protein to stromal cells leads to pre-metastatic niche formation[194]. Breast cancer cells EVs were found to contain miR-122 which could remodel metabolism to exacerbate metastasis[195]. VEGF-containing EVs can enhance EC glycolytic phenotype, inducing vascular permeability and cancer cells trans-endothelial migration[196] or promoting chemoresistance[197]. By increasing glycolysis and reprograming myeloid cells to an immunosuppressive phenotype, pancreatic ductal adenocarcinoma EVs could create an immunosuppressive background favoring tumor progression[198].
EVs can be involved in directional cell movement through tissues[199]. Distant spread can arise in two steps. The first one concerns local tumor cell dissemination where epithelial cell migrate through TME at the front of the tumor through generation of membrane protusions (invadopodia) and basal lamina break-in[200]. The second involves vascular disruption to allow tumor cells hematogenous spread. Once in the circulation, tumor cells migrate and must found a premetastatic niche where they can settle then proliferate.
To initiate both process, CRC cells will recruit then educate stromal cells to induce CAFs, tumor-associated macrophages with the immune-suppressive M2 phenotype, and endothelial cells that promote tumor angiogenesis[147]. CXCR4, present in HT29 EVs may also contribute to stromal cells recruitment[201]. CRC cells can induce CAF generation by EVs transfer of TGF-β[202] promoting also two CAFs distinct phenotypes, i.e., proliferative or invasive, by reprogramming their proteome[171]. Concerning macrophages, mutant p53 CRC cells are able to reprogram them into M2 phenotype through EVs miR-1246 transfer[203].
In both steps, loss of epithelial characteristics in favor of mesenchymal-like phenotype through epithelial to mesenchymal transition (EMT) process is involved[140,204]. During the local movement phase, stromal cells support EMT induction in tumor cells through stromal EVs. CAFs EVs can induce EMT in CRC cells by transfer of miR-92a-3p that promotes beta-catenin ubiquitination then degradation[205]. Similarly, EVs mediated transfer of miR-21 from CAFs to CRC cells increases their metastatic potential[166]. Aside CAFs, M2 macrophages can induce CRC cell migration through EVs cotransfer of miR21-5p and miR-155-5p[206]. M2 cells can also secrete Wnt-containing EVs to induce CRC stem cell activity that is involved in metastasis development[120]. This EMT transition is largely influenced by EVs matrixins transfer. Cotransfer of claudin 7 and MMP14 induces MMP2 and MMP9 recruitment that enhance invasiveness[207].
By EVs release, tumor cells can themselves induce up-or down-regulation of EMT-related genes in neighboring tumor cells, leading to distant invasion and/or migration[208]. EVs EMT inducers such as caveolin-1, HIF1α, beta-catenin, TNFa, TGF-β transfer can result in directional tumor cell migration[199,209] by either regulating ECM composition[210] or driving fibroblast differentiation into myofibroblast[211].
An important characteristic of tumor cells relies on their capacity to colonize preferentially specific organs (organotropic metastasis) that is often determined by anatomic aspects. Indeed, an important subset of CRCs will develop through distant metastasis, mostly to the liver. CRC capacenenity to colonize liver is primarily due to the hepatic portal system that drains the colon and by the facilitating defenestrated architecture of liver sinusoid endothelium[212]. Nevertheless, a crosstalk between CRC circulating cells and hepatocytes through bidirectional EVs transfer is also mandatory. It is now well accepted that primary tumor educates metastatic microenvironment, commonly defined as the “premetastatic niche,” allowing circulating tumor cells (CTC) to find a suitable environment in which they can settle then proliferate. Such niche generation is characterized by local tissue inflammation, immune suppression, stromal cell activation, and ECM remodeling[213]. EVs proteins or miRNAs have been shown to be involved in establishing this niche[167]. EVs can modify ECM to support circulating CRC cells adhesion by increasing fibronectin deposits within the liver[214]. Such ECM modifications increase CRC cell adhesion, promoting mesenchymal-to-epithelial transition (MET), and enabling liver metastasis colonization. EVs miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis[169] while EVs miR-21 through toll like receptor (TLR) 7/IL-6 axis in macrophages pathway as well as EVs miR-203 seem to induce an inflammatory niche that can potentiate liver metastasis[215,216]. EVs derived from CRC cell lines are involved in the modulation of the innate immune response, which is considered as a central step in the formation of the metastatic niche. Circulating EVs miRNAs after internalization by target cells can also act as ligands of TLRs[217].
Like in primary tumors, cancer cell EVs can reprogram resident cells to promote metastatic niche achievement and attract newly released CTCs. For example, in the niche, gastric cancer cells drive epidermal growth factor receptor (EGFR) EVs transfer to liver stromal cells that upregulate HGF expression through miR-26a/b downregulation inducing CTC attraction and further metastatic proliferation[218].
Angiogenesis is important for tumor proliferation and distant metastasis. Endothelial cells (ECs) can uptake via the endocytic pathway EVs from various origins[219]. Uptake of tumor-derived exosomes by normal endothelial cells activates angiogenic signaling pathways in endothelial cells and stimulates new vessel formation[67,68,220]. Once internalized, EVs are immediately directed to the perinuclear zone and actin filaments enriched area. When tubules are formed, EVs move to cell periphery and enter advanced pseudopods[221]. After complete remodeling, adjacent ECs probably transport EVs to neighboring ECs and to other cells in the TME[222].
In hypoxic conditions, tumor cells can secrete angiogenic factors, such as VEGF-A, inducing ECs migration and tumor angiogenesis. Higher levels of circulating proangiogenic basic bFGF originating from CRC cells have been detected in the serum of CRC patients[223]. EVs are also released by hypoxic CRC cells. Wnt4 enriched EVs increased β-catenin nuclear translocation in ECs enhancing angiogenesis and tumor growth[224]. It holds the same for Wnt5a[225] and Wnt5b whose increased expression in CRC cells correlates with aggressiveness. Caco-2 cells, one of the mostly used human CRC cell lines, secrete Wnt5b containing EVs that stimulates cell migration and proliferation of A549 cells[210]. Mutations in adenomatous polyposis coli (APC) gene are common in CRC patients and are associated with the deregulation in Wnt signaling. Restoration of APC expression in CRC SW480 cells induces DKK4 release through EVs, a mechanism restoring Wnt signaling pathway that may be lost during CRC progression[226]. In CRC ascites, EVs released by CRC tumor cells have been shown to carry proangiogenic proteins like Plexin B2 and tetraspanin[227]. Interestingly, CRC cell lines (HCT116 and DLD-1) secrete EVS that carry high levels of tissue factor, which is involved in blood coagulation, but is also a known modulator of angiogenesis and metastasis processes[228]. Aside proteins, EVs miRs have also been involved in angiogenesis induction[229], miR-183-5p was first found to be highly expressed in CRC cell-derived EVs, which triggers a marked increase in the proliferation, migration and tube formation abilities of HMEC-1 cells by targeting FOXO1[230]. CRC-derived miR-1229 containing EVs, by inhibiting HPIK2 expression, promote through VEGF pathway activation HUVECs tubulogenesis, transfection with exomiR-1229 inhibitor anta-miR-1229 significantly suppressing tube formation[231]. EVs from 5-FU-resistant CRC cells promoted angiogenesis through dipeptidyl peptidase IV, a potent inducer of this angiogenesis[232].
TAMs were also proven to be beneficial for angiogenesis. M2 macrophages were positively correlated with microvessel density of pancreatic ductal adenocarcinoma tissues. M2 macrophage-derived EVs could promote mouse aortic ECs angiogenesis in vitro and subcutaneous tumors growth in vivo, increasing vascular density in mice[233].
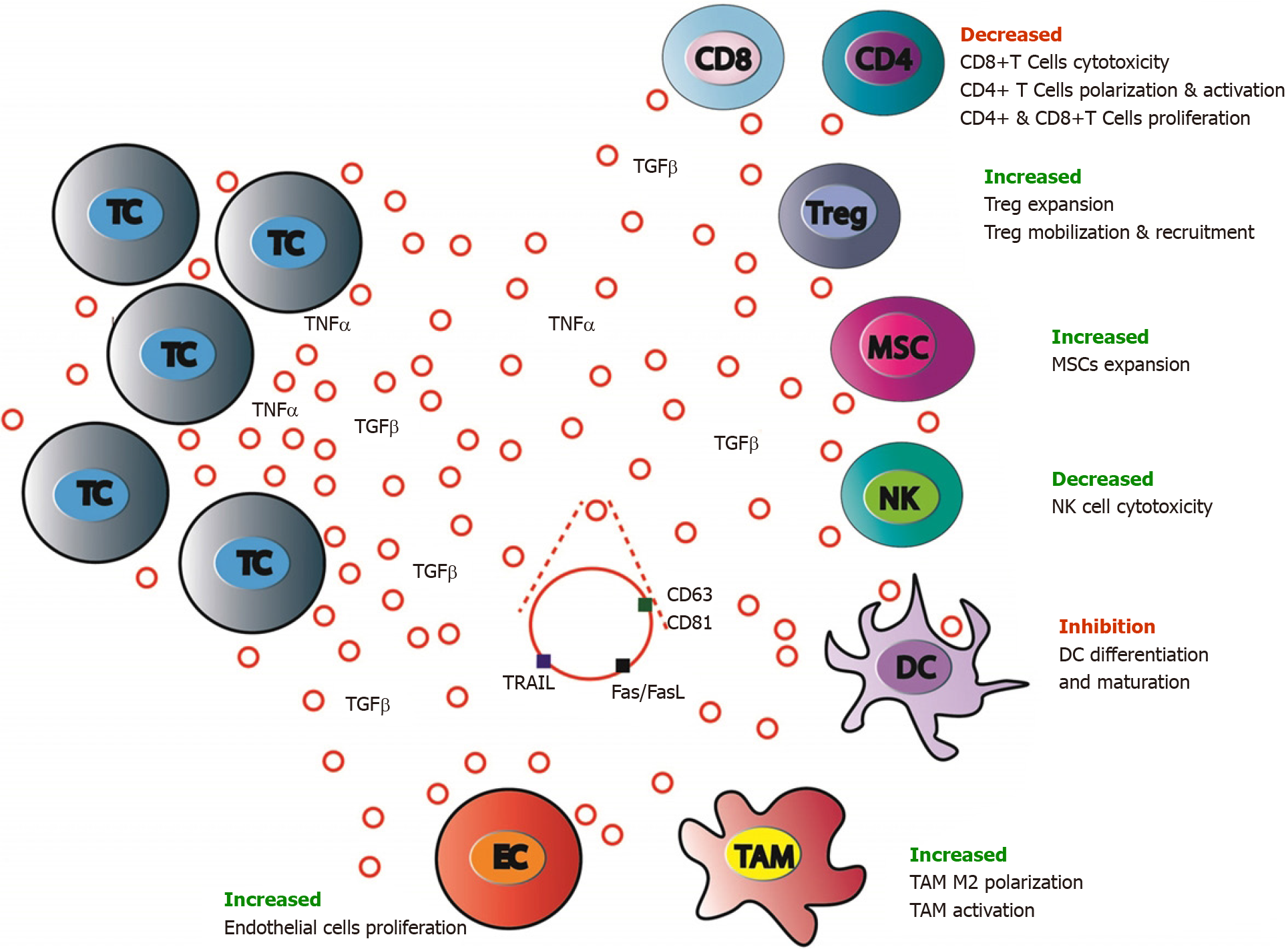
While tumor cell dissemination seems to be an early event of tumorigenesis, metastasis development ability is strongly associated with immune evasion. It seems that in CRC, the immune system influences tumor heterogeneity and sculpts clonal evolution. Tumor clones development is linked to the intra-metastatic immune microenvironment via an immune editing process[234].
CRC EVs induce recruitment to the pre-metastatic niche of suppressive immune cells, such as TAMs, tumor-associated neutrophils, Tregs leading to a strong inhibition of the antitumor response and facilitating CRC growth[235]. Specifically, it has been shown that TAMs can stimulate CRC growth by altering ECM remodeling, TME composition, tumor metabolism and angiogenesis[187]. CRC-derived EVs are involved these processes. CRC cells TGF-β EVs transfer to T cells can induce cell repro
Despite improvement and diversification of therapeutics for CRC patients (surgery, targeted therapy, radiotherapy and chemotherapy) and the emergence of new drugs during the last years, resistance to treatment still exists and remains one of the deadlocks for patients with an advanced CRC for whom medicines no longer work[242]. Today, administration of FOLFOX, a combination of folinic acid, 5- FU and oxaliplatin (OXA), is one of the most widely used chemotherapeutic regimens for treating CRC but these treatments generate serious systemic side effects and have an impact on the patients quality of life. More recently, the use of targeted drugs (for example bevacizumab, cetuximab, regorafenib ...) allow improvement of metastatic CRC survival times but malignant tumors drug resistance still persist[243].
Resistance to conventional chemotherapy: Aside classical mechanisms of resistance to 5-FU and OXA such as impaired drug inflow or efflux, drug inactivation, or single nucleotide polymorphisms of fluoropyrimidine or platinum targets, EVs generated by CRC cells have been reported to play a critical role in resistance to treatments[244]. Cancer stemness acquisition could be a possible feature that induces chemoresistance in CRC[245]. Wnt activity may reflect stem cell features. EVs-mediated Wnt secretion by CAFs is able to induce CRC reprogramming into CSCs then potentiate CRC resistance to chemotherapy[246]. In addition, CAFs release of H19 EVs also potentiated cancer stem cell resistance to OXA. LncRNA H19 was highly expressed in CAFs and upregulated in EVs. H19 activated the Wnt/β-catenin signaling pathway and potentiated drug resistance of CSCs[247]. The role of CAFs in exporting EVs that will confer chemoresistance to CRC cells is significant as it was reported that CAFs Evs can activate CRC cells ERK/AKT pathway inducing a protective effect to OXA[162]. CAFs can export urothelial carcinoma-associated 1 (UCA1), a lncRNA with three exons that has been found to display oncogenic functions in various types of cancer[248]. In CRC, UCA1 was found to be associated with resistance to cetuximab and 5-FU[249,250]. UCA1 suppresses miRNA-204-5p expression[251] that induces drug resistance. miR-196b-5p promotes CRC cells chemoresistance to 5-FU by targeting SOCS1 and SOCS3 negative regulators of STAT3 signaling pathway, resulting in global activation of STAT3 signaling[157]. Interestingly, UCA1 and miR-196b-5p are highly expressed in CRC patients EVs as compared to healthy control subjects and may represent interesting CRC biomarkers (Figure 5).
Resistance to targeted therapies: Cetuximab or panitumumab, that target the extracellular domain of EGFR preventing downstream activation of the MAPK or mTOR pathways, increases survival times in CRC patients[252]. Nevertheless, a subset of mutations involving either BRAF or PIK3 and amplifications of MET or HER2 induce resistance to these monoclonal antibodies (Mab) therapy[253]. Cetuximab CRC-resistant EVs have been shown to restrict the PI3K negative regulator PTEN in CRC cells[254] through UCA1 overexpression[250]. Aside EVs nucleic acids or proteins inhibition of EGFR-driven cellular process in the recipient cell, EGFR positive EVs could bind anti-EGFR mAbs reducing mAb bioavailability. Such mechanism has been described for anti VEGFA mAb bevacizumab in metastatic and lung cancers. VEGFA positive EVs neutralize bevacizumab inducing cancer cell chemotherapeutic escape[255].
Being able to quantify and use EVs as relevant biological markers may improve CRC screening in the future. Nowadays, CRC is currently detected by different methods. Colonoscopy is widely used in clinical practice, which is regarded as the gold standard for detecting CRC. However, it has several limitations such as invasive nature, high cost and bothering bowel preparation[256]. Aside this invasive procedure, non-invasive screening tests such as iterative fecal occult blood testing (FOBT)[257] or plasma carcinoembryonic antigen (CEA) quantification have also been used. Unfortunately, both are of limited value mainly because poor sensitivity and specificity[258,259] urging the need to find new methods aimed to quickly, easily and robustly diagnose and monitor CRC. This is where EVs can certainly play an important role.
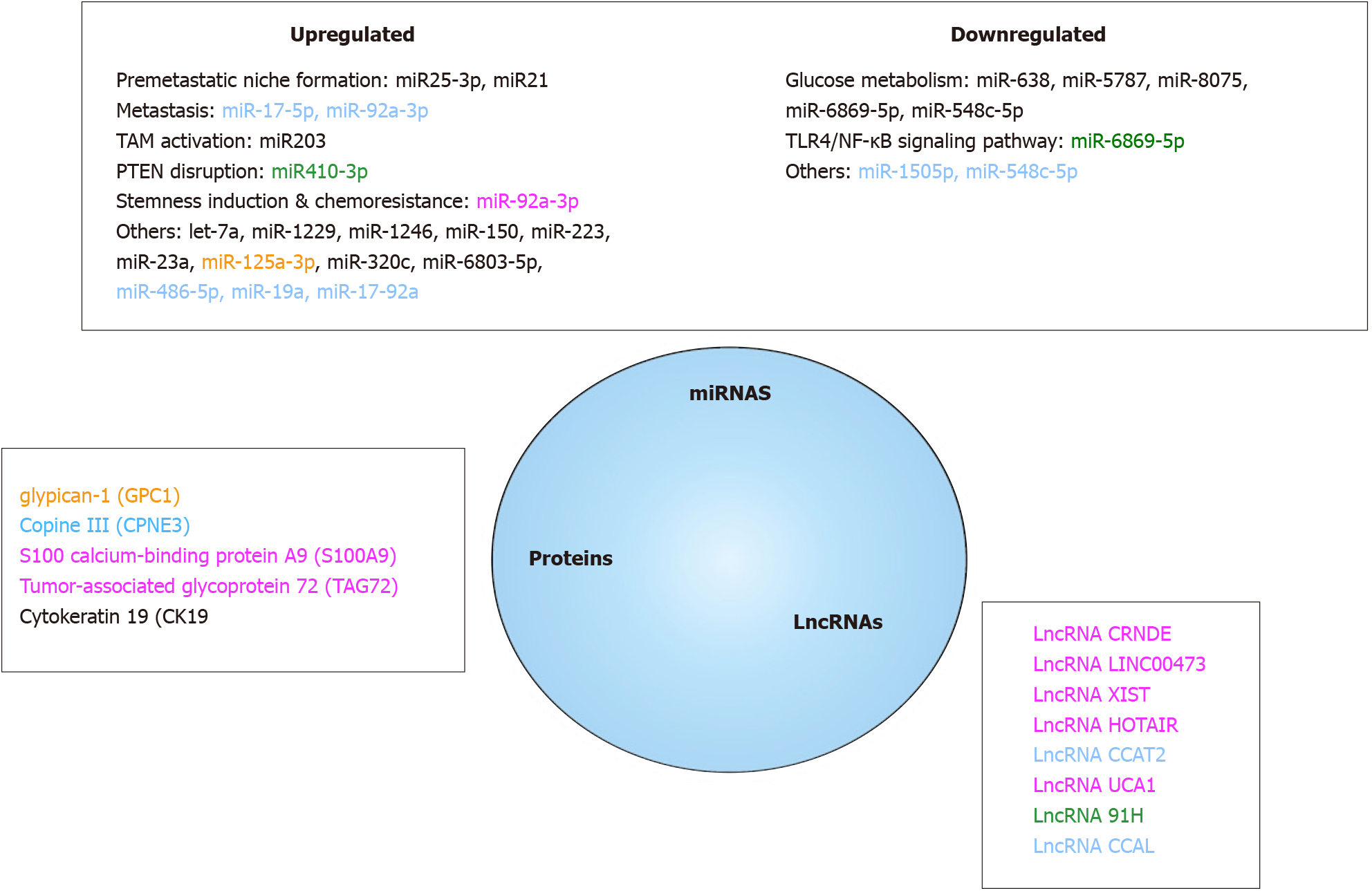
EVs can be detected in many biological fluids of patients, such as blood, urine, CSF and saliva[13] and can now be easily isolated[260] even though a universal standardized and widely accepted method for isolating then analyzing EVs is still mandatory[244]. Thanks to their lipid bilayers, EVs are stable in circulation and protected from degradation of serum ribonucleases and DNases[261]. As several miRNAs, lncRNAs and proteins are differently expressed in EVs originating from tumor and normal cells, they are potential sources of biomarkers and become a promising field in CRC diagnosis (Figure 6).
EVs miRNAs as relevant CRC biological markers: EVs miRs have been regularly involved in CRC development holding promise that their quantification in plasma or serum could serve as relevant CRC biomarkers. Some of them, that have been associated to specific events in CRC natural history, have been found in blood of CRC patients[262]. Among them, miR-25-3p[169] and miR-21[216], both promoting pre-metastatic niche formation by respectively inducing vascular permeability and macrophages differentiation towards a pro-inflammatory phenotype, and miR-203 that induces TAM activation[215], have been reported to be highly expressed in plasma CRC patients EVs and related to a poor prognosis. Recently, miR-410-3p was found highly enriched in hypoxic CRC-derived EVs in a HIF1α or HIF2α-dependent manner. miR-410-3p decreases PTEN in recipient cancer cells thus activating PI3/Akt axis and leading to tumor progression. miR-410-3p levels were positively associated with poor prognosis of CRC[141]. Nevertheless, while several specific miRNAs panels have been found in EVs from CRC patients, only a few have yet been clinically validated[263]. A panel of 7 miRNAs (let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a) was first validated by qRT-PCR, indicating that it may be a suitable biomarker to detect CRC[264]. Among this, miR-23a, miR-1246 and miR-21 are highly interesting as all three display high specificity and sensibility[262]. If both miR-23a and miR-1246 are positive and both CA19-9 and CEA negative, one can say that it is probably an early stage CRC[265]. In addition, miR-125a-3p and miR-320c were found to be significantly increased in EVs of early-stage CRC patients, combination of miR-125a-3P and CEA improving drastically the screening power for early-stage CRCs[266]. Another interesting work showed that miR-6803-5p was significantly increased in serum samples from CRC patients and correlated to a poor prognosis as compared to healthy subjects[267]. While associated increased levels of both miR-17-5p and miR-92a-3p levels may serve as an early indicator of liver metastases[268], EVs overexpression of miR-486-5p, miR-19a, miR-17-92a correlate with CRC recurrence[269,270]. Last, increased expression of EVs miRs that can be released by CAFs can be also an early indicator of chemotherapy resistance. High expression of miR-92a-3p activates Wnt/β-catenin pathway and inhibits mitochondrial apoptosis by directly inhibiting FBXW7 and MOAP1, contributing to cell stemness, EMT, metastasis and 5-FU resistance in CRC[205].
On the opposite, aside plasma EVs miRs increased levels, down-regulation of some miRNAs could be predictive factors of CRC. Five EVs miRNAs (miR-638, miR-5787, miR-8075, miR-6869-5p and miR-548c-5p) were decreased among CRC patients. These miRNAs may be involved in the development and progression of CRC by regulating glucose metabolism. Besides, in this study, 2 miRNAs (miR-486-5p and miR-3180-5p) have been shown to be significantly increased[271], results that were further confirmed[269]. Low levels of tumor suppressor miR-6869-5p that targets TLR4/NF-κB signaling pathway inhibiting proliferation and promoting CRC cells apoptosis have been reported in CRC patients serum EVs[272]. More recently, decreased expression of miR-1505p[273] and miR-548c-5p[274] were both associated to CRC poor prognosis.
LncRNAs as interesting CRC markers: LncRNAs, non-coding RNAs greater than 200 nucleotides, were once considered as junk DNA and transcriptional noise but emerging evidences demonstrate that they are evolutionarily conserved and that their strongly regulated expression plays critical roles in regulating gene expression[275]. As they can be differentially expressed in blood EVs of CRC patients, they could be new interesting biomarkers[276]. LncRNAs have been involved in CRC initiation and progression. Colorectal cancer-associated lncRNA (CCAL) seems to be a key regulator of CRC progression[277] and it was reported that CCAL promotes OXA resistance of CRC cells[278]. It has been also demonstrated that both down-regulation of lncRNA UCA1 and up-regulation of circRNA homeodomain interacting protein kinase 3 is found in CRC patients EVs. UCA1 LncRNAs, upregulated in CRC biopsies and downregulated in serum EVs, serves as a miR143 sponge that modulate MYO6 expression[279]. Six lncRNAs (LNCV6_116109, LNCV6_98390, LNCV6_38772, LNCV_108266, LNCV6_84003, and LNCV6_98602) are significantly up-regulated in patients with CRC as compared to healthy individuals[280]. High serum EVs expression of lncRNA 91H have been associated to CRC poor prognosis[281] and an increase of growth arrest-specific 5 and colon cancer-associated transcript 2 (CCAT2) lncRNAs in CRC patients have also been reported[282]. Interestingly, CCAT2 lncRNA levels were significantly decreased after surgery and removal of the tumor[283]. Finally, several lncRNAs have been associated to treatment resistance[284]. HOTAIR[285], XIST[286] and LINC00473[287] lncRNAs have been found to confer 5-FU resistance through respective miR-218 and miR-203a-3p, miR15a and miR-152 regulations[288,289]. LncRNA CRNDE induces CRC OXA resistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling and miR 136 sponging[290,291].
EVs proteins as a source of cancer biomarkers: Finally, aside nucleic acids, EVs proteins could also be measured to diagnose CRC as they may differ between healthy and CRC individuals. A primary study has shown that 36 proteins were upregulated and 22 proteins downregulated in CRC patients EVs compared to normal volunteers EVs. Moreover, upregulation of these proteins was associated with a pretumorigenic microenvironment for metastasis and on the opposite, downregulation was associated with tumor growth and cell survival[292]. Several studies have identified a number of proteins that can be considered as potential biomarkers. For example, among them, glypican-1[293,294] was suggested to be a specific diagnosis marker because it is highly expressed in CRC patient EVs and normalized after surgery. Identically, EVs lower expression of Copine III, a protein highly expressed in CRC tumors, was associated to better survival[295]. Additionally, S100 calcium-binding protein A9 (S100A9) levels were noticeably higher in plasma EVs of CRC relapse patients than those in tumor resection patients[296]. S100A9 has been related to CRC worsening as its overexpression could enhance TME CRC cells stemness. High levels of cytokeratin 19, CA125, and tumor-associated glycoprotein 72 (TAG72) have been quantified in CRC patients plasma EVs[297]. Interestingly, TAG72 protein overexpression was found to contribute to CRC patients chemoresistance to 5-FU.
The emergence of quantitative measurements that will be simple, inexpensive, easily performed and non-invasive for the patient is strongly mandatory. Analysis of EVs content (miRNAs, lncRNAs and proteins) may allow early diagnosing CRC and even predicting its relapse, metastasis and potential chemotherapy resistance.
EVs have been shown to be a source of patient’s resistance to chemotherapy. It is mandatory to explore new therapeutic possibilities aimed to both suppress tumor progression and reduce EVs-related drug resistance.
EVs uptake and biogenesis inhibitions: The first possibility to treat cancer would be to target EVs by inhibiting EVs uptake[298]. Indeed, EVs endocytosis is an active process but a rather complex one leading its inhibition a new therapeutic perspective but a very difficult one to achieve. Many studies have found molecules that could inhibit EVs internalization. Heparin can inhibit in a dose-dependent manner EVs absorption through direct action on heparan sulfate proteoglycans which themselves play a role EVs endocytosis[299]. Cytochalasin D that inhibits phagocytosis and other endocytosis mechanisms through an inhibitory effect of actin polymerization has been shown to inhibit EVs uptake[300]. Inhibition of EVs internalization by Methyl-β-cyclodextrin (MβCD) in glioblastoma cells has been reported[301]. MβCD depletes cholesterol from natural membranes and decreases EVs uptake by interfering with lipid rafts stability. Another molecule, dynamin, already described as an inhibitor of endocytosis, has been shown to interfere with EVs uptake in cancer[302]. Nevertheless, the large repertoire of mechanisms involved in EVs uptake in cancer impairs the overall efficiency of these molecules. A recent study showed that antibodies targeting CD9 and CD63 tetraspanins stimulate EVs macrophages phagocytose inhibiting cancer EVs-mediated communication[303]. However, such antibodies do not only target cancer EVs but also “physiological” CD9 and CD63 EVs. The role of these specific EVs being not yet known, additional studies must be carried out to know the viability of such method.
One other possibility of EVs targeting would be to inhibit EVs biogenesis. Inhibiting EVs biogenesis also involves complex issues, primarily due to the large number of proteins that are concerned in this cellular process. However many pharmacological agents have been found and seem promising. Fluidity of cell plasma membrane is fundamental during membrane lipid bilayer re-organization and thus EVs formation. During EVs biogenesis, ceramide regulate EVs production[24]. Ceramide synthesis required an ubiquitous enzyme, neutral sphingomyelinase 2 (nSMase2) that can be specifically targeted by GW4869 inhibiting cancer cells EVs release in a dose-dependent manner[304] and consequently limiting miRNAs hematogenous release[305]. On the opposite, nSMase2 overexpression increases miRNAs quantity in blood[306]. The link between nSmase2 and EVs has been shown in breast cancer aggressiveness[307]. GW4869 therapeutic effects have been observed on murine melanoma. GW4869-induced B16BL6-derived EVs secretion inhibition decreased B16BL6 cells proliferation and increased apoptosis-related proteins. Treatment of GW4869-treated cells with B16BL6-derived EVs restore their proliferation[308]. As GW4869 seems to be promising, imipramine which is a tricyclic anti-depressant is also a source of interest because of its inhibitory activity on acid sphingomyelinase (aSMase) that catalyzes sphingomyelin hydrolysis to ceramide[309]. Thus, imipramine is reported to prevent the translocation of aSMase, inhibiting EVs secretion. So, both GW4869 and imipramine can stop the production of ceramide
TSG101 is a protein involved on endosomes trafficking and exosomes biogenesis[310]. In CRC cells that express Wnt5b, knockdown of TSG-101 generates Wnt5b EVs downregulation decreasing Wnt5b-driven cell proliferation suggesting TSG101 as a potential therapeutic target in cancer[311].
EVs release inhibition: A third possibility to target EVs is to limit or inhibit their release by secreting cells.
A drug that inhibits EVs release is manumycin A, an antibiotic which is a selective and strong inhibitor of Ras farnesyltransferases. Farnesyltransferase inhibitors inhibit Ras activity and therefore EVs release[312]. Aside Ras proteins figure Rab proteins that are also modulators of EVs biogenesis[7]. Rab2b, Rab5a, Rab9a, Rab27a and Rab27b impacts in EVs release have been studied, the two latter playing also a role in EVs docking and exocytosis[29]. Knockdown of Rab27a decreased EVs-release amount[313] and Rab27a inhibition reduced tumor growth and lowered metastatic cells dissemination[314,315]. Gold nanoparticles conjugated with anti-sense RAB27a oligonucleotides to mute Rab27a generate 80% inhibition of EVs release in breast cancer[316]. Plectin enables EVs secretion in pancreatic cancer. Downregulation of plectin in pancreatic cancer cells reduced EVs release in the same way Rab27a and Rab27b knockdowns do suggesting that combining both mechanisms could be a therapeutic combination that enables greater results[317].
As plasma membrane fluidity is important for EVs shedding, drugs aimed at targeting either lipid rafts formation or cholesterol synthesis will interfere with EVs release. Lipid depletion results in EVs release reduction[318]. Pantethine, a pantothenic acid (vitamin B5) derivative is used as an intermediate in the production of co-enzyme A and it plays a role in the metabolism of lipids and reduction of total cholesterol levels. Panthetine inhibits by 80% cholesterol synthesis as well as fatty acid synthesis[319]. Panthetine has been shown to limit EVs release in systemic sclerosis[320]. Its use on chemoresistant breast cancer cells significantly reduced EVs release[321].
Actin and actin-regulating proteins are also strongly involved in EVs secretion. Invadopodia are cellular structures used by cancer cells to degrade extracellular matrix and invade. Because of high levels of actin, such structures are key sites for EVs release. Indeed, invadopodia inhibition limits EVs release[322]. Furthermore, knockdown of cortactin, that acts as an actin dynamics regulatory protein, decreased whereas its overexpression led to an increase of EVs release[323].
Rho-associated protein kinases (ROCK) are a family of serine-threonine kinases belonging to the PKA-G-C family and involved in cells shape and movement regulation, by acting on the cytoskeleton. Cytoskeleton organization as well as cellular contractility through activity on actin filaments is important features for EVs shedding. Y27632 is a commonly used ROCK competitive inhibitor which is able to compete with ATP at ROCK catalytic sites[324]. Y27632 causes a reduction in the release of EVs as well as a change in cell surface morphology[325] by sustaining activation of proteolytic enzymes, such as stathmin and calpain, that destabilized cell plasma membrane. Thus, Y27632 can be used alone or in combination with Calpeptin, the most studied calpain inhibitor[326]. Calpains, once activated through calcium binding, can activate different cellular processes including cell migration, cell invasion and EVs formation and release. Calpeptin has also been used alone to inhibit EVs release[327].
PEG-SMRwt-Clu, a drug derived from the secretion region of HIV-1 Nef protein, regulates exosomal pathway trafficking and seems promising. PEG-SMRwt-Clu was able to inhibit cell growth in breast cancer cell lines and more interesting to partially increase chemosensitivity. The use of PEG-SMRwt-Clu was also associated with a decrease in the number of released EVs[328].
Despite the current efforts and the number of EVs endocytosis, biogenesis and release inhibitors that are already available, inhibition of EVs is still a very complex issue because of the multifactorial nature of the different pathways involved in these processes. Nevertheless, EVs uptake, biogenesis or release inhibition remains a potential and interesting therapeutic cancer target in the near future.
EVs are major players in tumor progression via the transfer of cargo within them. One other possible way to cure CRC would be an EVs-based therapy that uses EVs as therapeutic vectors.
In very recent years, studies have mainly focused on the idea that EVs could be natural delivery vehicles to transport therapeutic drugs, antibodies or RNA to modify gene expression[329]. In the cancer field, it would be indeed a specific and effective therapy delivery method to specifically treat cancer cells. EVs are biocompatible and biodegradable and therefore, less toxic and immunogenic than other nanoparticular drug delivery systems such as liposomes or polymeric nanoparticles[330]. EVs have innate limited immunogenicity and cytotoxicity[331,332]. Moreover, drug stability is largely enhanced as EVs avoid drugs degradation by extracellular enzymes[333]. Thus EVs capacity to target tumor cells is 10 times higher than liposomes of a similar size. Such property is certainly linked to particular ligand-receptor interactions and to efficient endocytosis mechanisms linked to the EVs membrane lipid composition that contributes significantly to cellular adherence and internalization[334]. Last, EVs can penetrate through anatomical barriers[335,336] and their lipid composition protects them from reticuloendothelial system phagocytosis[244].
Several reports have demonstrated the potential of using EVs therapy and clinical trials are currently underway to find treatments that extend patient survival. Many kinds of EVs-based therapies have been shown to improve chemotherapy effectiveness. EVs have been used to deliver many kinds of drugs such as curcumin[337], paclitaxel[338] and doxorubicin[339]. While loading doxorubicin in EVs reduces cardiotoxicity[340], its packaging into EVs increases its efficacy when compared to free doxorubicin in cancer-bearing mice treatment. Inside EVs, doxorubicin has a better stability and will be even more collected within the tumor, significantly suppressing mice CRC growth and extending survival time[341]. EVs loaded with paclitaxel were tested in the treatment of multiple drug resistance cancers. Loaded exosomes can overcome drug efflux transporter adverse effect, decreasing metastasis growth when compared to controls[342].
EVs are also natural carriers of nucleic acids molecules and can be genetically engineered to deliver specific nucleic acid molecules such as miRNA[343], and more recently gene editing system CRISPR/Cas9[344]. EVs-based nucleic acid delivery in cancer treatment have shown promising therapeutic effects[38]. EGFR expressing cells can be targeted with GE11-positive exosomes loaded with microRNA let-7a, a tumor suppressor microRNA. The results showed an efficient delivery of exosomes cargo and consequent tumor growth inhibition[345].
EVs can also be used as a new type of tumor vaccine. Phase I clinical trials have shown that ascites EVs combination with granulocyte-macrophage colony stimulating factor induces a safe and effective response from specific anti-tumor cytotoxic T-cell in the treatment of advanced CRC[346]. EVs have also been explored as modulators of the immune response against tumor cells. Dendritic cells are antigen-presenting cells inducing immune responses. Dendritic cells have been shown to secrete antigen-presenting EVs that coexpress molecules of the major histocompatibility complex. Such exosomes activate specific cytotoxic T lymphocytes in vivo that can reduce or even suppress tumor growth[347]. EVs loading of anti-tumor peptides has also been used. A specific mutated form of survivin-T34A induces caspase activation leading to apoptosis. In vitro treatment of cancer cell lines with survivin-T34A EVs increased cell death[348].
Different cell-derived EVs may be home to specific cell types[7]. EVs derived from hypoxic tumor cells tend to be taken up by hypoxic tumor cells[349]. Different cells under different conditions determine EVs heterogeneity, generating huge and complex combinatorial possibilities. Thus, to better use EVs in cancer, engineering EVs with ligands that can specifically bind to targeted cancer cells is mandatory. Either EVs surface expression of receptor/ligand, antibody/ligand or microenvironment specific molecules can be used to specifically modify EVs. Recently, bioengineered EVs have been shown to be able to specifically bind to HER2/Neu by expressing designed ankyrin repeat proteins on their membrane surface[350]. Engineering both CD3 and EGFR expression on EVs membranes allows cross-linking of T cells with EGFR positive cancer cells enhancing antitumor immunity[351]. As hyaluronan has been evidenced in EVs[352], hyaluronidase engineered EVs have been shown to degrade tumor extracellular matrix and enhance the permeability of T cells and drugs within the tumor[353].
Using EVs as therapeutic vectors in cancer seems very promising and clinical trials are nowadays being carried out[354]. Unfortunately, no major breakthrough still occurs certainly because of the complexity to handle such new therapeutic methods in vivo. To accelerate their use in cancer patient treatment, there is also an urgent need to better understand both EVs biology and nature[298].
EVs exert a wide variety of biological functions, mainly via delivering signaling molecules that regulate a vast repertoire of cellular processes. Their role in cancer development is central as they participate through bidirectional signaling between cancer cells and TME cells to every step of CRC carcinogenesis up to metastatic dissemination. Their detection in a large variety of biological fluids represents the future of cancer detection, an easy and reproducible mean to identify specific biomarkers of diagnostic and prognostic relevance. Moreover, they also represent new targets for treatment as their inhibition could limit or stop cancer development. Additionally, as extracellular signaling molecules, they could be used as very specific nanovectors to transport conventional or innovative therapies to cancer cells of interest.
However, although pre-clinical data appear very promising, validation from large clinical trials are needed to support EVs use as either tumor biomarkers for monitoring cancer progression and driving treatment decisions or new vectors for specifically targeted treatments. Such data are mandatory to better understand EVs function in cancer progression and translate EVs use in clinical practice.
We are indebted to Mrs Pasquet for her fine work in the correction of the English text of our manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Oncology
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Haque N S-Editor: Zhang H L-Editor: A P-Editor: Liu JH
| 1. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3585] [Cited by in F6Publishing: 4282] [Article Influence: 713.7] [Reference Citation Analysis (1)] |
| 2. | Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1625] [Cited by in F6Publishing: 1466] [Article Influence: 244.3] [Reference Citation Analysis (0)] |
| 3. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12667] [Cited by in F6Publishing: 13879] [Article Influence: 3469.8] [Reference Citation Analysis (4)] |
| 4. | Herzig D, Hardiman K, Weiser M, You N, Paquette I, Feingold DL, Steele SR. The American Society of Colon and Rectal Surgeons Clinical Practice Guidelines for the Management of Inherited Polyposis Syndromes. Dis Colon Rectum. 2017;60:881-894. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 5. | Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-1437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4060] [Cited by in F6Publishing: 4984] [Article Influence: 498.4] [Reference Citation Analysis (0)] |
| 6. | Naito Y, Yoshioka Y, Yamamoto Y, Ochiya T. How cancer cells dictate their microenvironment: present roles of extracellular vesicles. Cell Mol Life Sci. 2017;74:697-713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 7. | Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3378] [Cited by in F6Publishing: 3537] [Article Influence: 353.7] [Reference Citation Analysis (0)] |
| 8. | Margolis L, Sadovsky Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019;17:e3000363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 301] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 9. | Witwer KW, Théry C. Extracellular vesicles or exosomes? J Extracell Vesicles. 2019;8:1648167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 320] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 10. | Shah R, Patel T, Freedman JE. Circulating Extracellular Vesicles in Human Disease. N Engl J Med. 2018;379:2180-2181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 11. | Rahbarghazi R, Jabbari N, Sani NA, Asghari R, Salimi L, Kalashani SA, Feghhi M, Etemadi T, Akbariazar E, Mahmoudi M, Rezaie J. Tumor-derived extracellular vesicles: reliable tools for Cancer diagnosis and clinical applications. Cell Commun Signal. 2019;17:73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 12. | Nazarenko I. Extracellular Vesicles: Recent Developments in Technology and Perspectives for Cancer Liquid Biopsy. Recent Results Cancer Res. 2020;215:319-344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4900] [Cited by in F6Publishing: 5527] [Article Influence: 502.5] [Reference Citation Analysis (0)] |
| 14. | Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers EM, Laitinen S, Lässer C, Lener T, Ligeti E, Linē A, Lipps G, Llorente A, Lötvall J, Manček-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-'t Hoen EN, Nyman TA, O'Driscoll L, Olivan M, Oliveira C, Pállinger É, Del Portillo HA, Reventós J, Rigau M, Rohde E, Sammar M, Sánchez-Madrid F, Santarém N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3453] [Cited by in F6Publishing: 3446] [Article Influence: 382.9] [Reference Citation Analysis (0)] |
| 15. | Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D'Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-'t Hoen EN, Noren Hooten N, O'Driscoll L, O'Grady T, O'Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6453] [Cited by in F6Publishing: 6146] [Article Influence: 1024.3] [Reference Citation Analysis (0)] |
| 16. | Meehan B, Rak J, Di Vizio D. Oncosomes - large and small: what are they, where they came from? J Extracell Vesicles. 2016;5:33109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 17. | Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhães A, Ferreira JA, Osório H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, Lyden D. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20:332-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 741] [Cited by in F6Publishing: 992] [Article Influence: 165.3] [Reference Citation Analysis (0)] |
| 18. | Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364-372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 786] [Cited by in F6Publishing: 937] [Article Influence: 104.1] [Reference Citation Analysis (0)] |
| 19. | Caruso S, Poon IKH. Apoptotic Cell-Derived Extracellular Vesicles: More Than Just Debris. Front Immunol. 2018;9:1486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 353] [Cited by in F6Publishing: 326] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 20. | Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem Sci. 2017;42:42-56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 300] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 21. | Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 956] [Cited by in F6Publishing: 1045] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 22. | Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 467] [Cited by in F6Publishing: 534] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 23. | Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1573] [Cited by in F6Publishing: 1466] [Article Influence: 244.3] [Reference Citation Analysis (0)] |
| 24. | Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244-1247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2186] [Cited by in F6Publishing: 2446] [Article Influence: 152.9] [Reference Citation Analysis (0)] |
| 25. | Falguières T, Luyet PP, Gruenberg J. Molecular assemblies and membrane domains in multivesicular endosome dynamics. Exp Cell Res. 2009;315:1567-1573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Scott CC, Gruenberg J. Ion flux and the function of endosomes and lysosomes: pH is just the start: the flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. Bioessays. 2011;33:103-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 27. | Anand S, Samuel M, Kumar S, Mathivanan S. Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim Biophys Acta Proteins Proteom. 2019;1867:140203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 163] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 28. | Guardia CM, Farías GG, Jia R, Pu J, Bonifacino JS. BORC Functions Upstream of Kinesins 1 and 3 to Coordinate Regional Movement of Lysosomes along Different Microtubule Tracks. Cell Rep. 2016;17:1950-1961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 29. | Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19-30; sup pp 1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1525] [Cited by in F6Publishing: 1703] [Article Influence: 113.5] [Reference Citation Analysis (0)] |
| 30. | Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24:35-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 219] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 31. | Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1605] [Cited by in F6Publishing: 1477] [Article Influence: 98.5] [Reference Citation Analysis (0)] |
| 32. | Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, Li M, Shi L, Pan C, Zhu D, Chen X, Hu G, Liu Y, Zhang CY, Zen K. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun. 2017;8:14041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 33. | Saunderson SC, Dunn AC, Crocker PR, McLellan AD. CD169 mediates the capture of exosomes in spleen and lymph node. Blood. 2014;123:208-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 256] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 34. | Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T, Takakura Y. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013;165:77-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 419] [Cited by in F6Publishing: 481] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 35. | Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1483] [Cited by in F6Publishing: 2098] [Article Influence: 419.6] [Reference Citation Analysis (0)] |
| 36. | Horibe S, Tanahashi T, Kawauchi S, Murakami Y, Rikitake Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer. 2018;18:47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 187] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 37. | Costa Verdera H, Gitz-Francois JJ, Schiffelers RM, Vader P. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J Control Release. 2017;266:100-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 279] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 38. | Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1167] [Cited by in F6Publishing: 1543] [Article Influence: 220.4] [Reference Citation Analysis (0)] |
| 39. | Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50:1025-1034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 174] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 40. | Muntasell A, Berger AC, Roche PA. T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO J. 2007;26:4263-4272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 182] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 41. | Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968-E977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2360] [Cited by in F6Publishing: 2234] [Article Influence: 279.3] [Reference Citation Analysis (0)] |
| 42. | Jin Y, Chen K, Wang Z, Wang Y, Liu J, Lin L, Shao Y, Gao L, Yin H, Cui C, Tan Z, Liu L, Zhao C, Zhang G, Jia R, Du L, Chen Y, Liu R, Xu J, Hu X. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer. 2016;16:753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 43. | Zebrowska A, Skowronek A, Wojakowska A, Widlak P, Pietrowska M. Metabolome of Exosomes: Focus on Vesicles Released by Cancer Cells and Present in Human Body Fluids. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 44. | Rontogianni S, Synadaki E, Li B, Liefaard MC, Lips EH, Wesseling J, Wu W, Altelaar M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019;2:325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 45. | Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, Meckes DG Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget. 2016;7:86999-87015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 176] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 46. | Jeannin P, Chaze T, Giai Gianetto Q, Matondo M, Gout O, Gessain A, Afonso PV. Proteomic analysis of plasma extracellular vesicles reveals mitochondrial stress upon HTLV-1 infection. Sci Rep. 2018;8:5170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 47. | Turchinovich A, Drapkina O, Tonevitsky A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front Immunol. 2019;10:202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 164] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 48. | Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143-1149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 741] [Cited by in F6Publishing: 762] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 49. | Miranda KC, Bond DT, Levin JZ, Adiconis X, Sivachenko A, Russ C, Brown D, Nusbaum C, Russo LM. Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PLoS One. 2014;9:e96094. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 50. | Phinney DG, Di Giuseppe M, Njah J, Sala E, Shiva S, St Croix CM, Stolz DB, Watkins SC, Di YP, Leikauf GD, Kolls J, Riches DW, Deiuliis G, Kaminski N, Boregowda SV, McKenna DH, Ortiz LA. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat Commun. 2015;6:8472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 519] [Cited by in F6Publishing: 630] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 51. | Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A, Kalluri R. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869-3875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 646] [Cited by in F6Publishing: 725] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 52. | Antonyak MA, Wilson KF, Cerione RA. R(h)oads to microvesicles. Small GTPases. 2012;3:219-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 53. | Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 176] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 54. | Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Grønborg M, Möbius W, Rhee J, Barr FA, Simons M. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 545] [Cited by in F6Publishing: 583] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 55. | Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128:3171-3176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 267] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 56. | Verweij FJ, Bebelman MP, Jimenez CR, Garcia-Vallejo JJ, Janssen H, Neefjes J, Knol JC, de Goeij-de Haas R, Piersma SR, Baglio SR, Verhage M, Middeldorp JM, Zomer A, van Rheenen J, Coppolino MG, Hurbain I, Raposo G, Smit MJ, Toonen RFG, van Niel G, Pegtel DM. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. 2018;217:1129-1142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 180] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 57. | Vietri M, Radulovic M, Stenmark H. The many functions of ESCRTs. Nat Rev Mol Cell Biol. 2020;21:25-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 457] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 58. | Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, Lochnit G, Preissner KT, Zöller M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668-1678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 453] [Cited by in F6Publishing: 492] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 59. | Guix FX, Sannerud R, Berditchevski F, Arranz AM, Horré K, Snellinx A, Thathiah A, Saido T, Saito T, Rajesh S, Overduin M, Kumar-Singh S, Radaelli E, Corthout N, Colombelli J, Tosi S, Munck S, Salas IH, Annaert W, De Strooper B. Tetraspanin 6: a pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol Neurodegener. 2017;12:25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 60. | Pollet H, Conrard L, Cloos AS, Tyteca D. Plasma Membrane Lipid Domains as Platforms for Vesicle Biogenesis and Shedding? Biomolecules. 2018;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 61. | Mazurov D, Barbashova L, Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013;280:1200-1213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 62. | Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, Vázquez J, Yáñez-Mó M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288:11649-11661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 344] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 63. | Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 2011;30:2115-2129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 64. | Jang SC, Crescitelli R, Cvjetkovic A, Belgrano V, Olofsson Bagge R, Sundfeldt K, Ochiya T, Kalluri R, Lötvall J. Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. J Extracell Vesicles. 2019;8:1635420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 65. | Gatti P, Ilamathi HS, Todkar K, Germain M. Mitochondria Targeted Viral Replication and Survival Strategies-Prospective on SARS-CoV-2. Front Pharmacol. 2020;11:578599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 66. | Pasquier J, Guerrouahen BS, Al Thawadi H, Ghiabi P, Maleki M, Abu-Kaoud N, Jacob A, Mirshahi M, Galas L, Rafii S, Le Foll F, Rafii A. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med. 2013;11:94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 322] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 67. | Pasquier J, Thawadi HA, Ghiabi P, Abu-Kaoud N, Maleki M, Guerrouahen BS, Vidal F, Courderc B, Ferron G, Martinez A, Al Sulaiti H, Gupta R, Rafii S, Rafii A. Microparticles mediated cross-talk between tumoral and endothelial cells promote the constitution of a pro-metastatic vascular niche through Arf6 up regulation. Cancer Microenviron. 2014;7:41-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 68. | Al Thawadi H, Abu-Kaoud N, Al Farsi H, Hoarau-Véchot J, Rafii S, Rafii A, Pasquier J. VE-cadherin cleavage by ovarian cancer microparticles induces β-catenin phosphorylation in endothelial cells. Oncotarget. 2016;7:5289-5305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 69. | Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361-374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1080] [Cited by in F6Publishing: 1190] [Article Influence: 170.0] [Reference Citation Analysis (0)] |
| 70. | Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012;109:4146-4151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 416] [Cited by in F6Publishing: 474] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 71. | Wang Q, Lu Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat Commun. 2017;8:709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 72. | Griffiths RE, Kupzig S, Cogan N, Mankelow TJ, Betin VM, Trakarnsanga K, Massey EJ, Lane JD, Parsons SF, Anstee DJ. Maturing reticulocytes internalize plasma membrane in glycophorin A-containing vesicles that fuse with autophagosomes before exocytosis. Blood. 2012;119:6296-6306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 133] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 73. | Xu X, Lai Y, Hua ZC. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep. 2019;39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 223] [Cited by in F6Publishing: 412] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 74. | Turiák L, Misják P, Szabó TG, Aradi B, Pálóczi K, Ozohanics O, Drahos L, Kittel A, Falus A, Buzás EI, Vékey K. Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice. J Proteomics. 2011;74:2025-2033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 75. | Pasquier J, Thomas B, Hoarau-Véchot J, Odeh T, Robay A, Chidiac O, Dargham SR, Turjoman R, Halama A, Fakhro K, Menzies R, Jayyousi A, Zirie M, Al Suwaidi J, Rafii A, Malik RA, Talal T, Abi Khalil C. Circulating microparticles in acute diabetic Charcot foot exhibit a high content of inflammatory cytokines, and support monocyte-to-osteoclast cell induction. Sci Rep. 2017;7:16450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 76. | Puhka M, Takatalo M, Nordberg ME, Valkonen S, Nandania J, Aatonen M, Yliperttula M, Laitinen S, Velagapudi V, Mirtti T, Kallioniemi O, Rannikko A, Siljander PR, Af Hällström TM. Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes. Theranostics. 2017;7:3824-3841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 77. | Bruschi M, Ravera S, Santucci L, Candiano G, Bartolucci M, Calzia D, Lavarello C, Inglese E, Petretto A, Ghiggeri G, Panfoli I. The human urinary exosome as a potential metabolic effector cargo. Expert Rev Proteomics. 2015;12:425-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 78. | Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60:9-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 367] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 79. | Ronquist KG, Ek B, Morrell J, Stavreus-Evers A, Ström Holst B, Humblot P, Ronquist G, Larsson A. Prostasomes from four different species are able to produce extracellular adenosine triphosphate (ATP). Biochim Biophys Acta. 2013;1830:4604-4610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 80. | Kohnhorst CL, Kyoung M, Jeon M, Schmitt DL, Kennedy EL, Ramirez J, Bracey SM, Luu BT, Russell SJ, An S. Identification of a multienzyme complex for glucose metabolism in living cells. J Biol Chem. 2017;292:9191-9203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 81. | Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A, Nagrath D. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 505] [Cited by in F6Publishing: 625] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 82. | Clement E, Lazar I, Attané C, Carrié L, Dauvillier S, Ducoux-Petit M, Esteve D, Menneteau T, Moutahir M, Le Gonidec S, Dalle S, Valet P, Burlet-Schiltz O, Muller C, Nieto L. Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. EMBO J. 2020;39:e102525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 152] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 83. | Gudbergsson JM, Johnsen KB, Skov MN, Duroux M. Systematic review of factors influencing extracellular vesicle yield from cell cultures. Cytotechnology. 2016;68:579-592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 84. | Palviainen M, Saari H, Kärkkäinen O, Pekkinen J, Auriola S, Yliperttula M, Puhka M, Hanhineva K, Siljander PR. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J Extracell Vesicles. 2019;8:1596669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 85. | Yang E, Wang X, Gong Z, Yu M, Wu H, Zhang D. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct Target Ther. 2020;5:242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 168] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 86. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8246] [Cited by in F6Publishing: 9027] [Article Influence: 531.0] [Reference Citation Analysis (0)] |
| 87. | Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131-139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 560] [Cited by in F6Publishing: 611] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 88. | Ridder K, Sevko A, Heide J, Dams M, Rupp AK, Macas J, Starmann J, Tjwa M, Plate KH, Sültmann H, Altevogt P, Umansky V, Momma S. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology. 2015;4:e1008371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 89. | Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, Wurdinger T, Pegtel DM, van Rheenen J. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046-1057. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 560] [Cited by in F6Publishing: 621] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 90. | Lv Z, Wei Y, Wang D, Zhang CY, Zen K, Li L. Argonaute 2 in cell-secreted microvesicles guides the function of secreted miRNAs in recipient cells. PLoS One. 2014;9:e103599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 91. | Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen HW, Baldini N, Pegtel DM. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6:127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 443] [Cited by in F6Publishing: 534] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 92. | Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell TK, Kahn CR. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 188] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 93. | O'Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21:585-606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 494] [Cited by in F6Publishing: 869] [Article Influence: 217.3] [Reference Citation Analysis (0)] |
| 94. | Wojtuszkiewicz A, Schuurhuis GJ, Kessler FL, Piersma SR, Knol JC, Pham TV, Jansen G, Musters RJ, van Meerloo J, Assaraf YG, Kaspers GJ, Zweegman S, Cloos J, Jimenez CR. Exosomes Secreted by Apoptosis-Resistant Acute Myeloid Leukemia (AML) Blasts Harbor Regulatory Network Proteins Potentially Involved in Antagonism of Apoptosis. Mol Cell Proteomics. 2016;15:1281-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 95. | Sousa D, Lima RT, Vasconcelos MH. Intercellular Transfer of Cancer Drug Resistance Traits by Extracellular Vesicles. Trends Mol Med. 2015;21:595-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 96. | Pasquier J, Galas L, Boulangé-Lecomte C, Rioult D, Bultelle F, Magal P, Webb G, Le Foll F. Different modalities of intercellular membrane exchanges mediate cell-to-cell p-glycoprotein transfers in MCF-7 breast cancer cells. J Biol Chem. 2012;287:7374-7387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 97. | Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766-769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 997] [Cited by in F6Publishing: 1136] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 98. | Chang X, Fang L, Bai J, Wang Z. Characteristics and Changes of DNA in Extracellular Vesicles. DNA Cell Biol. 2020;39:1486-1493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 99. | Fischer S, Cornils K, Speiseder T, Badbaran A, Reimer R, Indenbirken D, Grundhoff A, Brunswig-Spickenheier B, Alawi M, Lange C. Indication of Horizontal DNA Gene Transfer by Extracellular Vesicles. PLoS One. 2016;11:e0163665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 100. | Vagner T, Spinelli C, Minciacchi VR, Balaj L, Zandian M, Conley A, Zijlstra A, Freeman MR, Demichelis F, De S, Posadas EM, Tanaka H, Di Vizio D. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7:1505403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 239] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 101. | Lázaro-Ibáñez E, Sanz-Garcia A, Visakorpi T, Escobedo-Lucea C, Siljander P, Ayuso-Sacido A, Yliperttula M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate. 2014;74:1379-1390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 193] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 102. | Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, Davis G, Kumar T, Katz M, Overman MJ, Foretova L, Fabianova E, Holcatova I, Janout V, Meric-Bernstam F, Gascoyne P, Wistuba I, Varadhachary G, Brennan P, Hanash S, Li D, Maitra A, Alvarez H. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 323] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 103. | Hur JY, Kim HJ, Lee JS, Choi CM, Lee JC, Jung MK, Pack CG, Lee KY. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol Cancer. 2018;17:15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 104. | Liang Y, Qiao L, Peng X, Cui Z, Yin Y, Liao H, Jiang M, Li L. The chemokine receptor CCR1 is identified in mast cell-derived exosomes. Am J Transl Res. 2018;10:352-367. [PubMed] [Cited in This Article: ] |
| 105. | Guescini M, Guidolin D, Vallorani L, Casadei L, Gioacchini AM, Tibollo P, Battistelli M, Falcieri E, Battistin L, Agnati LF, Stocchi V. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. 2010;316:1977-1984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 106. | Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna). 2010;117:1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 428] [Cited by in F6Publishing: 458] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 107. | Ye W, Tang X, Yang Z, Liu C, Zhang X, Jin J, Lyu J. Plasma-derived exosomes contribute to inflammation via the TLR9-NF-κB pathway in chronic heart failure patients. Mol Immunol. 2017;87:114-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 108. | Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly L, Galkin A, Thakur BK, Soplop N, Uryu K, Hoshino A, Norton L, Bonafé M, Cricca M, Gasparre G, Lyden D, Bromberg J. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A. 2017;114:E9066-E9075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 449] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 109. | van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 512] [Cited by in F6Publishing: 531] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 110. | Lin XP, Almqvist N, Telemo E. Human small intestinal epithelial cells constitutively express the key elements for antigen processing and the production of exosomes. Blood Cells Mol Dis. 2005;35:122-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 111. | Mallegol J, van Niel G, Heyman M. Phenotypic and functional characterization of intestinal epithelial exosomes. Blood Cells Mol Dis. 2005;35:11-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 112. | Marzesco AM, Janich P, Wilsch-Bräuninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118:2849-2858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 354] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 113. | Lucchetti D, Calapà F, Palmieri V, Fanali C, Carbone F, Papa A, De Maria R, De Spirito M, Sgambato A. Differentiation Affects the Release of Exosomes from Colon Cancer Cells and Their Ability to Modulate the Behavior of Recipient Cells. Am J Pathol. 2017;187:1633-1647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 114. | Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420-1424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 551] [Cited by in F6Publishing: 558] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 115. | Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012; 143: 1518-1529. e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 475] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 116. | Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, Edison, Aliyev J, Wu Y, Bunte R, Williams BO, Rossant J, Virshup DM. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206-2215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 257] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 117. | Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895-902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1150] [Cited by in F6Publishing: 1088] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 118. | Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, Ivanov JA, Fu YY, Takashima S, Hua G, Martin ML, O'Rourke KP, Lo YH, Mokry M, Romera-Hernandez M, Cupedo T, Dow L, Nieuwenhuis EE, Shroyer NF, Liu C, Kolesnick R, van den Brink MRM, Hanash AM. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 756] [Cited by in F6Publishing: 723] [Article Influence: 80.3] [Reference Citation Analysis (1)] |
| 119. | Zhang L, Wrana JL. The emerging role of exosomes in Wnt secretion and transport. Curr Opin Genet Dev. 2014;27:14-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 120. | Saha S, Aranda E, Hayakawa Y, Bhanja P, Atay S, Brodin NP, Li J, Asfaha S, Liu L, Tailor Y, Zhang J, Godwin AK, Tome WA, Wang TC, Guha C, Pollard JW. Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nat Commun. 2016;7:13096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 179] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 121. | Das S, Yu S, Sakamori R, Vedula P, Feng Q, Flores J, Hoffman A, Fu J, Stypulkowski E, Rodriguez A, Dobrowolski R, Harada A, Hsu W, Bonder EM, Verzi MP, Gao N. Rab8a vesicles regulate Wnt ligand delivery and Paneth cell maturation at the intestinal stem cell niche. Development. 2015;142:2147-2162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 122. | Abdelhamed S, Butler JT, Doron B, Halse A, Nemecek E, Wilmarth PA, Marks DL, Chang BH, Horton T, Kurre P. Extracellular vesicles impose quiescence on residual hematopoietic stem cells in the leukemic niche. EMBO Rep. 2019;20:e47546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 123. | Macia L, Nanan R, Hosseini-Beheshti E, Grau GE. Host- and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int J Mol Sci. 2019;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 124. | Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, Hutchins E, Mu J, Deng Z, Luo C, Sundaram K, Sriwastva MK, Zhang L, Hsieh M, Reiman R, Haribabu B, Yan J, Jala VR, Miller DM, Van Keuren-Jensen K, Merchant ML, McClain CJ, Park JW, Egilmez NK, Zhang HG. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018; 24: 637-652. e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 437] [Cited by in F6Publishing: 431] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 125. | Barteneva NS, Baiken Y, Fasler-Kan E, Alibek K, Wang S, Maltsev N, Ponomarev ED, Sautbayeva Z, Kauanova S, Moore A, Beglinger C, Vorobjev IA. Extracellular vesicles in gastrointestinal cancer in conjunction with microbiota: On the border of Kingdoms. Biochim Biophys Acta Rev Cancer. 2017;1868:372-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 126. | Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, Ceroni A, Babayan SA, Blaxter M, Ivens A, Maizels RM. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 508] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 127. | Wang L, Li Z, Shen J, Liu Z, Liang J, Wu X, Sun X, Wu Z. Exosome-like vesicles derived by Schistosoma japonicum adult worms mediates M1 type immune- activity of macrophage. Parasitol Res. 2015;114:1865-1873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 128. | McDaniel K, Correa R, Zhou T, Johnson C, Francis H, Glaser S, Venter J, Alpini G, Meng F. Functional role of microvesicles in gastrointestinal malignancies. Ann Transl Med. 2013;1:4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 7] [Reference Citation Analysis (0)] |
| 129. | Yang Y, Xu C, Wu D, Wang Z, Wu P, Li L, Huang J, Qiu F. γδ T Cells: Crosstalk Between Microbiota, Chronic Inflammation, and Colorectal Cancer. Front Immunol. 2018;9:1483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 130. | Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1410] [Cited by in F6Publishing: 1524] [Article Influence: 152.4] [Reference Citation Analysis (0)] |
| 131. | Na YR, Stakenborg M, Seok SH, Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat Rev Gastroenterol Hepatol. 2019;16:531-543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 424] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 132. | Wong WY, Lee MM, Chan BD, Kam RK, Zhang G, Lu AP, Tai WC. Proteomic profiling of dextran sulfate sodium induced acute ulcerative colitis mice serum exosomes and their immunomodulatory impact on macrophages. Proteomics. 2016;16:1131-1145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 133. | Mitsuhashi S, Feldbrügge L, Csizmadia E, Mitsuhashi M, Robson SC, Moss AC. Luminal Extracellular Vesicles (EVs) in Inflammatory Bowel Disease (IBD) Exhibit Proinflammatory Effects on Epithelial Cells and Macrophages. Inflamm Bowel Dis. 2016;22:1587-1595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 134. | Geem D, Harusato A, Flannigan K, Denning TL. Harnessing regulatory T cells for the treatment of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1409-1418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 135. | Aiello S, Rocchetta F, Longaretti L, Faravelli S, Todeschini M, Cassis L, Pezzuto F, Tomasoni S, Azzollini N, Mister M, Mele C, Conti S, Breno M, Remuzzi G, Noris M, Benigni A. Extracellular vesicles derived from T regulatory cells suppress T cell proliferation and prolong allograft survival. Sci Rep. 2017;7:11518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 136. | Fais S, Venturi G, Gatenby B. Microenvironmental acidosis in carcinogenesis and metastases: new strategies in prevention and therapy. Cancer Metastasis Rev. 2014;33:1095-1108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 137. | Ban JJ, Lee M, Im W, Kim M. Low pH increases the yield of exosome isolation. Biochem Biophys Res Commun. 2015;461:76-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 138. | Logozzi M, Mizzoni D, Angelini DF, Di Raimo R, Falchi M, Battistini L, Fais S. Microenvironmental pH and Exosome Levels Interplay in Human Cancer Cell Lines of Different Histotypes. Cancers (Basel). 2018;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 123] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 139. | King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 650] [Cited by in F6Publishing: 732] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 140. | Huang Z, Yang M, Li Y, Yang F, Feng Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Transfer Wnt4 to Normoxic Cells to Elicit a Prometastatic Phenotype. Int J Biol Sci. 2018;14:2094-2102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 141. | Hu X, Mu Y, Liu J, Mu X, Gao F, Chen L, Wu H, Liu W, Zhao Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Transfer miR-410-3p to Regulate Tumor Progression. J Cancer. 2020;11:4724-4735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 142. | Iyer DN, Sin WY, Ng L. Linking stemness with colorectal cancer initiation, progression, and therapy. World J Stem Cells. 2019;11:519-534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 143. | Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 252] [Cited by in F6Publishing: 275] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 144. | Zhou Y, Xia L, Wang H, Oyang L, Su M, Liu Q, Lin J, Tan S, Tian Y, Liao Q, Cao D. Cancer stem cells in progression of colorectal cancer. Oncotarget. 2018;9:33403-33415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 145. | Munro MJ, Wickremesekera SK, Peng L, Tan ST, Itinteang T. Cancer stem cells in colorectal cancer: a review. J Clin Pathol. 2018;71:110-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 185] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 146. | Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1047] [Cited by in F6Publishing: 1030] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 147. | Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14:306-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 532] [Cited by in F6Publishing: 493] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 148. | Gao W, Chen L, Ma Z, Du Z, Zhao Z, Hu Z, Li Q. Isolation and phenotypic characterization of colorectal cancer stem cells with organ-specific metastatic potential. Gastroenterology 2013; 145: 636-46. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 149. | Humphries HN, Wickremesekera SK, Marsh RW, Brasch HD, Mehrotra S, Tan ST, Itinteang T. Characterization of Cancer Stem Cells in Colon Adenocarcinoma Metastasis to the Liver. Front Surg. 2017;4:76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 150. | Francescangeli F, Contavalli P, De Angelis ML, Careccia S, Signore M, Haas TL, Salaris F, Baiocchi M, Boe A, Giuliani A, Tcheremenskaia O, Pagliuca A, Guardiola O, Minchiotti G, Colace L, Ciardi A, D'Andrea V, La Torre F, Medema J, De Maria R, Zeuner A. A pre-existing population of ZEB2+ quiescent cells with stemness and mesenchymal features dictate chemoresistance in colorectal cancer. J Exp Clin Cancer Res. 2020;39:2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 151. | Sreepadmanabh M, Toley BJ. Investigations into the cancer stem cell niche using in-vitro 3-D tumor models and microfluidics. Biotechnol Adv. 2018;36:1094-1110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 152. | Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 883] [Cited by in F6Publishing: 1021] [Article Influence: 127.6] [Reference Citation Analysis (0)] |
| 153. | Prieto-Vila M, Takahashi RU, Usuba W, Kohama I, Ochiya T. Drug Resistance Driven by Cancer Stem Cells and Their Niche. Int J Mol Sci. 2017;18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 325] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 154. | Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847-856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1088] [Cited by in F6Publishing: 1163] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 155. | Stik G, Crequit S, Petit L, Durant J, Charbord P, Jaffredo T, Durand C. Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells. J Cell Biol. 2017;216:2217-2230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 156. | Ciardiello C, Leone A, Budillon A. The Crosstalk between Cancer Stem Cells and Microenvironment Is Critical for Solid Tumor Progression: The Significant Contribution of Extracellular Vesicles. Stem Cells Int. 2018;2018:6392198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 157. | Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y, Liang Y, Cao L, Li X, Li R, Sun L, Liu Q, Wu J, Zhou K, Zeng J. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807-49823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 158. | Chen S, Huang EH. The colon cancer stem cell microenvironment holds keys to future cancer therapy. J Gastrointest Surg. 2014;18:1040-1048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 159. | Vautrot V, Chanteloup G, Elmallah M, Cordonnier M, Aubin F, Garrido C, Gobbo J. Exosomal miRNA: Small Molecules, Big Impact in Colorectal Cancer. J Oncol. 2019;2019:8585276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 160. | Mu W, Rana S, Zöller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15:875-887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 192] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 161. | Paolillo M, Schinelli S. Integrins and Exosomes, a Dangerous Liaison in Cancer Progression. Cancers (Basel). 2017;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 162. | Bhome R, Goh RW, Bullock MD, Pillar N, Thirdborough SM, Mellone M, Mirnezami R, Galea D, Veselkov K, Gu Q, Underwood TJ, Primrose JN, De Wever O, Shomron N, Sayan AE, Mirnezami AH. Exosomal microRNAs derived from colorectal cancer-associated fibroblasts: role in driving cancer progression. Aging (Albany NY). 2017;9:2666-2694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 163. | Bhome R, Mellone M, Emo K, Thomas GJ, Sayan AE, Mirnezami AH. The Colorectal Cancer Microenvironment: Strategies for Studying the Role of Cancer-Associated Fibroblasts. Methods Mol Biol. 2018;1765:87-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 164. | Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene. 2015;34:5857-5868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 158] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 165. | Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, Xue Y, Xu T, Zhu HJ, Simpson RJ. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672-1686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 258] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 166. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1350] [Cited by in F6Publishing: 1417] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 167. | Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol Ther. 2016;17:1062-1069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 168. | Ragusa M, Statello L, Maugeri M, Barbagallo C, Passanisi R, Alhamdani MS, Li Destri G, Cappellani A, Barbagallo D, Scalia M, Valadi H, Hoheisel JD, Di Pietro C, Purrello M. Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: biomolecular, genetic and translational implications. Oncoscience. 2014;1:132-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 169. | Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, Hu J, Zhu X, Yang W, Liao W, Li G, Ding Y, Liang L. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 569] [Article Influence: 94.8] [Reference Citation Analysis (0)] |
| 170. | Lugini L, Valtieri M, Federici C, Cecchetti S, Meschini S, Condello M, Signore M, Fais S. Exosomes from human colorectal cancer induce a tumor-like behavior in colonic mesenchymal stromal cells. Oncotarget. 2016;7:50086-50098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 171. | Rai A, Greening DW, Chen M, Xu R, Ji H, Simpson RJ. Exosomes Derived from Human Primary and Metastatic Colorectal Cancer Cells Contribute to Functional Heterogeneity of Activated Fibroblasts by Reprogramming Their Proteome. Proteomics. 2019;19:e1800148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 172. | Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, Mala T, Sjo OH, Bakka A, Moberg I, Fetveit T, Mathisen Ø, Husby A, Sandvik O, Nesbakken A, Thiis-Evensen E, Lothe RA. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 173. | Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, Zhang B, Coffey RJ, Patton JG. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 263] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 174. | Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 415] [Cited by in F6Publishing: 433] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 175. | Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 388] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 176. | Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, Yan J, Miller DM, Zhang HG. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 317] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 177. | Yoshii S, Hayashi Y, Iijima H, Inoue T, Kimura K, Sakatani A, Nagai K, Fujinaga T, Hiyama S, Kodama T, Shinzaki S, Tsujii Y, Watabe K, Takehara T. Exosomal microRNAs derived from colon cancer cells promote tumor progression by suppressing fibroblast TP53 expression. Cancer Sci. 2019;110:2396-2407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 178. | Soldevilla B, Rodríguez M, San Millán C, García V, Fernández-Periañez R, Gil-Calderón B, Martín P, García-Grande A, Silva J, Bonilla F, Domínguez G. Tumor-derived exosomes are enriched in ΔNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014;23:467-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 179. | Fang M, Li Y, Huang K, Qi S, Zhang J, Zgodzinski W, Majewski M, Wallner G, Gozdz S, Macek P, Kowalik A, Pasiarski M, Grywalska E, Vatan L, Nagarsheth N, Li W, Zhao L, Kryczek I, Wang G, Wang Z, Zou W, Wang L. IL33 Promotes Colon Cancer Cell Stemness via JNK Activation and Macrophage Recruitment. Cancer Res. 2017;77:2735-2745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 180. | Lehuédé C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM. Metabolic Plasticity as a Determinant of Tumor Growth and Metastasis. Cancer Res. 2016;76:5201-5208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 180] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 181. | Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41:211-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1910] [Cited by in F6Publishing: 2554] [Article Influence: 319.3] [Reference Citation Analysis (1)] |
| 182. | Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71:6921-6925. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 628] [Cited by in F6Publishing: 724] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 183. | Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 776] [Cited by in F6Publishing: 861] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 184. | Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678-3684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 736] [Cited by in F6Publishing: 852] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 185. | Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, Lisanti MP. Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol. 2011;43:1045-1051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 186. | Bonuccelli G, Whitaker-Menezes D, Castello-Cros R, Pavlides S, Pestell RG, Fatatis A, Witkiewicz AK, Vander Heiden MG, Migneco G, Chiavarina B, Frank PG, Capozza F, Flomenberg N, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. The reverse Warburg effect: glycolysis inhibitors prevent the tumor promoting effects of caveolin-1 deficient cancer associated fibroblasts. Cell Cycle. 2010;9:1960-1971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 171] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 187. | Zhong X, Chen B, Yang Z. The Role of Tumor-Associated Macrophages in Colorectal Carcinoma Progression. Cell Physiol Biochem. 2018;45:356-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 188. | Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, Bescós C, Di Croce L, Benitah SA. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 668] [Cited by in F6Publishing: 856] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 189. | Rohlenova K, Veys K, Miranda-Santos I, De Bock K, Carmeliet P. Endothelial Cell Metabolism in Health and Disease. Trends Cell Biol. 2018;28:224-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 143] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 190. | Zhu L, Zhao Q, Yang T, Ding W, Zhao Y. Cellular metabolism and macrophage functional polarization. Int Rev Immunol. 2015;34:82-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 241] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 191. | Wang C, Xu J, Yuan D, Bai Y, Pan Y, Zhang J, Shao C. Exosomes carrying ALDOA and ALDH3A1 from irradiated lung cancer cells enhance migration and invasion of recipients by accelerating glycolysis. Mol Cell Biochem. 2020;469:77-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 192. | Li Y, Zhao Z, Liu W, Li X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl Biochem Biotechnol. 2020;191:1084-1099. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 193. | Shu S, Yang Y, Allen CL, Maguire O, Minderman H, Sen A, Ciesielski MJ, Collins KA, Bush PJ, Singh P, Wang X, Morgan M, Qu J, Bankert RB, Whiteside TL, Wu Y, Ernstoff MS. Metabolic reprogramming of stromal fibroblasts by melanoma exosome microRNA favours a pre-metastatic microenvironment. Sci Rep. 2018;8:12905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 194. | Dai J, Escara-Wilke J, Keller JM, Jung Y, Taichman RS, Pienta KJ, Keller ET. Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. J Exp Med. 2019;216:2883-2899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 195. | Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, Somlo G, Palomares M, Li Z, Tremblay JR, Tsuyada A, Sun G, Reid MA, Wu X, Swiderski P, Ren X, Shi Y, Kong M, Zhong W, Chen Y, Wang SE. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 687] [Cited by in F6Publishing: 813] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 196. | Hida K, Maishi N, Annan DA, Hida Y. Contribution of Tumor Endothelial Cells in Cancer Progression. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 197. | Wang B, Wang X, Hou D, Huang Q, Zhan W, Chen C, Liu J, You R, Xie J, Chen P, Huang H. Exosomes derived from acute myeloid leukemia cells promote chemoresistance by enhancing glycolysis-mediated vascular remodeling. J Cell Physiol. 2019;234:10602-10614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 198. | Basso D, Gnatta E, Padoan A, Fogar P, Furlanello S, Aita A, Bozzato D, Zambon CF, Arrigoni G, Frasson C, Franchin C, Moz S, Brefort T, Laufer T, Navaglia F, Pedrazzoli S, Basso G, Plebani M. PDAC-derived exosomes enrich the microenvironment in MDSCs in a SMAD4-dependent manner through a new calcium related axis. Oncotarget. 2017;8:84928-84944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 199. | Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun. 2015;6:7164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 387] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 200. | Lafitte M, Lecointre C, Roche S. Roles of exosomes in metastatic colorectal cancer. Am J Physiol Cell Physiol. 2019;317:C869-C880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 201. | Wang X, Ding X, Nan L, Wang Y, Wang J, Yan Z, Zhang W, Sun J, Zhu W, Ni B, Dong S, Yu L. Investigation of the roles of exosomes in colorectal cancer liver metastasis. Oncol Rep. 2015;33:2445-2453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 202. | Wu J, Li H, Xie H, Wu X, Lan P. The malignant role of exosomes in the communication among colorectal cancer cell, macrophage and microbiome. Carcinogenesis. 2019;40:601-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 203. | Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 328] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 204. | Guo J, Wang B, Fu Z, Wei J, Lu W. Hypoxic Microenvironment Induces EMT and Upgrades Stem-Like Properties of Gastric Cancer Cells. Technol Cancer Res Treat. 2016;15:60-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 205. | Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, Liao WT, Ding YQ, Liang L. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 431] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 206. | Lan J, Sun L, Xu F, Liu L, Hu F, Song D, Hou Z, Wu W, Luo X, Wang J, Yuan X, Hu J, Wang G. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019;79:146-158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 402] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 207. | Philip R, Heiler S, Mu W, Büchler MW, Zöller M, Thuma F. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget. 2015;6:2046-2063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 208. | Greening DW, Gopal SK, Mathias RA, Liu L, Sheng J, Zhu HJ, Simpson RJ. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 156] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 209. | Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18:75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 696] [Cited by in F6Publishing: 752] [Article Influence: 150.4] [Reference Citation Analysis (0)] |
| 210. | Harada T, Yoshimura K, Yamashita O, Ueda K, Morikage N, Sawada Y, Hamano K. Focal Adhesion Kinase Promotes the Progression of Aortic Aneurysm by Modulating Macrophage Behavior. Arterioscler Thromb Vasc Biol. 2017;37:156-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 211. | Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621-9630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 544] [Cited by in F6Publishing: 621] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 212. | Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XH. Metastasis Organotropism: Redefining the Congenial Soil. Dev Cell. 2019;49:375-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 177] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 213. | Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, Bissell MJ, Cox TR, Giaccia AJ, Erler JT, Hiratsuka S, Ghajar CM, Lyden D. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1003] [Cited by in F6Publishing: 1108] [Article Influence: 158.3] [Reference Citation Analysis (0)] |
| 214. | Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816-826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1554] [Cited by in F6Publishing: 1823] [Article Influence: 202.6] [Reference Citation Analysis (0)] |
| 215. | Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, Iguchi T, Ito S, Eguchi H, Ochiya T, Yanaga K, Miyano S, Mimori K. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8:78598-78613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 216. | Shao Y, Chen T, Zheng X, Yang S, Xu K, Chen X, Xu F, Wang L, Shen Y, Wang T, Zhang M, Hu W, Ye C, Yu X, Shao J, Zheng S. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis. 2018;39:1368-1379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 151] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 217. | Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110-E2116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1064] [Cited by in F6Publishing: 1155] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 218. | Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Yang H, Li J, Ning T, Huang D, Li H, Zhang L, Ying G, Ba Y. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 356] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 219. | Huang A, Dong J, Li S, Wang C, Ding H, Li H, Su X, Ge X, Sun L, Bai C, Shen X, Fang T, Li J, Shao N. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci. 2015;11:961-969. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 220. | Whiteside TL. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem. 2016;74:103-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 393] [Cited by in F6Publishing: 492] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 221. | Mineo M, Garfield SH, Taverna S, Flugy A, De Leo G, Alessandro R, Kohn EC. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis. 2012;15:33-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 226] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 222. | Gonda A, Kabagwira J, Senthil GN, Wall NR. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol Cancer Res. 2019;17:337-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 162] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 223. | Jibiki N, Saito N, Kameoka S, Kobayashi M. Clinical significance of fibroblast growth factor (FGF) expression in colorectal cancer. Int Surg. 2014;99:493-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 224. | Huang Z, Feng Y. Exosomes Derived From Hypoxic Colorectal Cancer Cells Promote Angiogenesis Through Wnt4-Induced β-Catenin Signaling in Endothelial Cells. Oncol Res. 2017;25:651-661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 105] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 225. | Ekström EJ, Bergenfelz C, von Bülow V, Serifler F, Carlemalm E, Jönsson G, Andersson T, Leandersson K. WNT5A induces release of exosomes containing pro-angiogenic and immunosuppressive factors from malignant melanoma cells. Mol Cancer. 2014;13:88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 226. | Lim JW, Mathias RA, Kapp EA, Layton MJ, Faux MC, Burgess AW, Ji H, Simpson RJ. Restoration of full-length APC protein in SW480 colon cancer cells induces exosome-mediated secretion of DKK-4. Electrophoresis. 2012;33:1873-1880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 227. | Choi DS, Park JO, Jang SC, Yoon YJ, Jung JW, Choi DY, Kim JW, Kang JS, Park J, Hwang D, Lee KH, Park SH, Kim YK, Desiderio DM, Kim KP, Gho YS. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011;11:2745-2751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 228. | Yu JL, May L, Lhotak V, Shahrzad S, Shirasawa S, Weitz JI, Coomber BL, Mackman N, Rak JW. Oncogenic events regulate tissue factor expression in colorectal cancer cells: implications for tumor progression and angiogenesis. Blood. 2005;105:1734-1741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 388] [Cited by in F6Publishing: 401] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 229. | Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440-2448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 718] [Cited by in F6Publishing: 710] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 230. | Shang A, Wang X, Gu C, Liu W, Sun J, Zeng B, Chen C, Ji P, Wu J, Quan W, Yao Y, Wang W, Sun Z, Li D. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging (Albany NY). 2020;12:8352-8371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 231. | Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, Chen Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 232. | Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085-1099. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 381] [Cited by in F6Publishing: 407] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 233. | Yang Y, Guo Z, Chen W, Wang X, Cao M, Han X, Zhang K, Teng B, Cao J, Wu W, Cao P, Huang C, Qiu Z. M2 Macrophage-Derived Exosomes Promote Angiogenesis and Growth of Pancreatic Ductal Adenocarcinoma by Targeting E2F2. Mol Ther. 2021;29:1226-1238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 234. | Angelova M, Mlecnik B, Vasaturo A, Bindea G, Fredriksen T, Lafontaine L, Buttard B, Morgand E, Bruni D, Jouret-Mourin A, Hubert C, Kartheuser A, Humblet Y, Ceccarelli M, Syed N, Marincola FM, Bedognetti D, Van den Eynde M, Galon J. Evolution of Metastases in Space and Time under Immune Selection. Cell 2018; 175: 751-765. e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 236] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 235. | Colby JK, Jaoude J, Liu F, Shureiqi I. Oxygenated lipid signaling in tumor-associated macrophages-focus on colon cancer. Cancer Metastasis Rev. 2018;37:289-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 236. | Yamada N, Kuranaga Y, Kumazaki M, Shinohara H, Taniguchi K, Akao Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-β1-mediated suppression. Oncotarget. 2016;7:27033-27043. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 237. | Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, Song M, Bian Z, Wang Q, Ni S, Hu Y, Mao Y, Zhou L, Wang Y, Yu J, Du X, Hua D, Huang Z. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187-6199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 238. | Sun B, Zhou Y, Fang Y, Li Z, Gu X, Xiang J. Colorectal cancer exosomes induce lymphatic network remodeling in lymph nodes. Int J Cancer. 2019;145:1648-1659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 239. | Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4:e1027472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 162] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 240. | Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, Zaccheddu A, Colone M, Arancia G, Gentile M, Seregni E, Valenti R, Ballabio G, Belli F, Leo E, Parmiani G, Rivoltini L. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128:1796-1804. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 372] [Cited by in F6Publishing: 381] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 241. | Plebanek MP, Angeloni NL, Vinokour E, Li J, Henkin A, Martinez-Marin D, Filleur S, Bhowmick R, Henkin J, Miller SD, Ifergan I, Lee Y, Osman I, Thaxton CS, Volpert OV. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat Commun. 2017;8:1319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 242. | Van der Jeught K, Xu HC, Li YJ, Lu XB, Ji G. Drug resistance and new therapies in colorectal cancer. World J Gastroenterol. 2018;24:3834-3848. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 356] [Cited by in F6Publishing: 343] [Article Influence: 57.2] [Reference Citation Analysis (5)] |
| 243. | Geng F, Wang Z, Yin H, Yu J, Cao B. Molecular Targeted Drugs and Treatment of Colorectal Cancer: Recent Progress and Future Perspectives. Cancer Biother Radiopharm. 2017;32:149-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 244. | Mannavola F, Salerno T, Passarelli A, Tucci M, Internò V, Silvestris F. Revisiting the Role of Exosomes in Colorectal Cancer: Where Are We Now? Front Oncol. 2019;9:521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 245. | Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, Pickell K, Aguilar J, Lazetic S, Smith-Berdan S, Clarke MF, Hoey T, Lewicki J, Gurney AL. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3:e2428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 403] [Cited by in F6Publishing: 463] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 246. | Hu YB, Yan C, Mu L, Mi YL, Zhao H, Hu H, Li XL, Tao DD, Wu YQ, Gong JP, Qin JC. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene. 2019;38:1951-1965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 247. | Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932-3948. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 469] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 248. | Xue M, Chen W, Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J Cancer Res Clin Oncol. 2016;142:1407-1419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 249. | Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, Zhou L, Qi X, Huang S, Hua D, Xing C, Huang Z. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 291] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 250. | Yang YN, Zhang R, Du JW, Yuan HH, Li YJ, Wei XL, Du XX, Jiang SL, Han Y. Predictive role of UCA1-containing exosomes in cetuximab-resistant colorectal cancer. Cancer Cell Int. 2018;18:164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 251. | Wu H, Liang Y, Shen L. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol Open. 2016;5:563-570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 252. | Arnold D, Lueza B, Douillard JY, Peeters M, Lenz HJ, Venook A, Heinemann V, Van Cutsem E, Pignon JP, Tabernero J, Cervantes A, Ciardiello F. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28:1713-1729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 477] [Cited by in F6Publishing: 562] [Article Influence: 93.7] [Reference Citation Analysis (0)] |
| 253. | Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, Yu Q, Yuan X. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017;8:3980-4000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 254. | Zhang S, Zhang Y, Qu J, Che X, Fan Y, Hou K, Guo T, Deng G, Song N, Li C, Wan X, Qu X, Liu Y. Exosomes promote cetuximab resistance via the PTEN/Akt pathway in colon cancer cells. Braz J Med Biol Res. 2017;51:e6472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 255. | Simon T, Pinioti S, Schellenberger P, Rajeeve V, Wendler F, Cutillas PR, King A, Stebbing J, Giamas G. Shedding of bevacizumab in tumour cells-derived extracellular vesicles as a new therapeutic escape mechanism in glioblastoma. Mol Cancer. 2018;17:132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 256. | Lieberman D. Colorectal Cancer Screening With Colonoscopy. JAMA Intern Med. 2016;176:903-904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 257. | De Maio G, Rengucci C, Zoli W, Calistri D. Circulating and stool nucleic acid analysis for colorectal cancer diagnosis. World J Gastroenterol. 2014;20:957-967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 28] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 258. | Elsafi SH, Alqahtani NI, Zakary NY, Al Zahrani EM. The sensitivity, specificity, predictive values, and likelihood ratios of fecal occult blood test for the detection of colorectal cancer in hospital settings. Clin Exp Gastroenterol. 2015;8:279-284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 259. | Sur D, Colceriu M, Sur G, Floca E, Dascal L, Irimie A. Colorectal cancer: evolution of screening strategies. Med Pharm Rep. 2019;92:21-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 260. | Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018;118:1917-1950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 628] [Cited by in F6Publishing: 895] [Article Influence: 149.2] [Reference Citation Analysis (0)] |
| 261. | Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 765] [Cited by in F6Publishing: 741] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 262. | Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, Yuan J, Sun J, Dai L, Yang C, Li Z, Wang J, Zhong T. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020;476:13-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 263. | Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20:1272-1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 264. | Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 557] [Cited by in F6Publishing: 596] [Article Influence: 59.6] [Reference Citation Analysis (1)] |
| 265. | Karimi N, Ali Hosseinpour Feizi M, Safaralizadeh R, Hashemzadeh S, Baradaran B, Shokouhi B, Teimourian S. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J Chin Med Assoc. 2019;82:215-220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 266. | Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, Zhang Y, Huang X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7:4150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 267. | Yan S, Jiang Y, Liang C, Cheng M, Jin C, Duan Q, Xu D, Yang L, Zhang X, Ren B, Jin P. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J Cell Biochem. 2018;119:4113-4119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 268. | Fu F, Jiang W, Zhou L, Chen Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl Oncol. 2018;11:221-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 269. | Liu X, Chen X, Zeng K, Xu M, He B, Pan Y, Sun H, Pan B, Xu X, Xu T, Hu X, Wang S. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018;9:1037. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 270. | Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y, Shinden Y, Eguchi H, Yamamoto H, Doki Y, Mori M, Ochiya T, Mimori K. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275-281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 332] [Cited by in F6Publishing: 372] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 271. | Yan S, Han B, Gao S, Wang X, Wang Z, Wang F, Zhang J, Xu D, Sun B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget. 2017;8:60149-60158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 272. | Yan S, Liu G, Jin C, Wang Z, Duan Q, Xu J, Xu D. MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-κB signaling pathway in colorectal cancer. J Cell Physiol. 2018;233:6660-6668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 273. | Zou SL, Chen YL, Ge ZZ, Qu YY, Cao Y, Kang ZX. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark. 2019;26:69-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 274. | Peng ZY, Gu RH, Yan B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem. 2018;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 275. | Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3151] [Cited by in F6Publishing: 3450] [Article Influence: 265.4] [Reference Citation Analysis (0)] |
| 276. | Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551-85563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 235] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 277. | Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494-1504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 254] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 278. | Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, Kong F, Guan M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146:1700-1716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 136] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 279. | Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D, Biondi A, Cappellani A, Basile F, Di Pietro C, Purrello M, Ragusa M. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol Ther Nucleic Acids. 2018;12:229-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 280. | Hu D, Zhan Y, Zhu K, Bai M, Han J, Si Y, Zhang H, Kong D. Plasma Exosomal Long Non-Coding RNAs Serve as Biomarkers for Early Detection of Colorectal Cancer. Cell Physiol Biochem. 2018;51:2704-2715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 281. | Gao T, Liu X, He B, Nie Z, Zhu C, Zhang P, Wang S. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 282. | Liu L, Meng T, Yang XH, Sayim P, Lei C, Jin B, Ge L, Wang HJ. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22:283-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 283. | Wang L, Duan W, Yan S, Xie Y, Wang C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed Pharmacother. 2019;113:108758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 284. | Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell Oncol (Dordr). 2019;42:757-768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 285. | Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR Contributes to 5FU Resistance through Suppressing miR-218 and Activating NF-κB/TS Signaling in Colorectal Cancer. Mol Ther Nucleic Acids. 2020;20:879-880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 286. | Xiao Y, Yurievich UA, Yosypovych SV. Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget. 2017;8:83171-83182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 287. | Wang L, Zhang X, Sheng L, Qiu C, Luo R. LINC00473 promotes the Taxol resistance via miR-15a in colorectal cancer. Biosci Rep. 2018;38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 288. | Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a Prognostic Biomarker for the Proliferation and Chemoresistance of Colorectal Cancer via MiR-203a-3p-Mediated Wnt/ß-Catenin Signaling Pathway. Cell Physiol Biochem. 2018;46:1275-1285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 289. | Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, Liu Y. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 261] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 290. | Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 371] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 291. | Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 292. | Chen Y, Xie Y, Xu L, Zhan S, Xiao Y, Gao Y, Wu B, Ge W. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer. 2017;140:900-913. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 293. | Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z, Tang Y, Liu W, Zhu B, Wang L, Ren C. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med. 2017;21:838-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 294. | Campanella C, Rappa F, Sciumè C, Marino Gammazza A, Barone R, Bucchieri F, David S, Curcurù G, Caruso Bavisotto C, Pitruzzella A, Geraci G, Modica G, Farina F, Zummo G, Fais S, Conway de Macario E, Macario AJ, Cappello F. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer. 2015;121:3230-3239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 109] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 295. | Sun B, Li Y, Zhou Y, Ng TK, Zhao C, Gan Q, Gu X, Xiang J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234:1416-1425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 296. | Wang Y, Yin K, Tian J, Xia X, Ma J, Tang X, Xu H, Wang S. Granulocytic Myeloid-Derived Suppressor Cells Promote the Stemness of Colorectal Cancer Cells through Exosomal S100A9. Adv Sci (Weinh). 2019;6:1901278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 297. | Xiao Y, Li Y, Yuan Y, Liu B, Pan S, Liu Q, Qi X, Zhou H, Dong W, Jia L. The potential of exosomes derived from colorectal cancer as a biomarker. Clin Chim Acta. 2019;490:186-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 298. | Bastos N, Ruivo CF, da Silva S, Melo SA. Exosomes in cancer: Use them or target them? Semin Cell Dev Biol. 2018;78:13-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 299. | Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A. 2013;110:17380-17385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 551] [Cited by in F6Publishing: 625] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 300. | Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1298] [Cited by in F6Publishing: 1682] [Article Influence: 168.2] [Reference Citation Analysis (0)] |
| 301. | Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Mörgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288:17713-17724. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 434] [Cited by in F6Publishing: 496] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 302. | Kawamoto T, Ohga N, Akiyama K, Hirata N, Kitahara S, Maishi N, Osawa T, Yamamoto K, Kondoh M, Shindoh M, Hida Y, Hida K. Tumor-derived microvesicles induce proangiogenic phenotype in endothelial cells via endocytosis. PLoS One. 2012;7:e34045. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 303. | Nishida-Aoki N, Tominaga N, Takeshita F, Sonoda H, Yoshioka Y, Ochiya T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol Ther. 2017;25:181-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 304. | Nakamura K, Sawada K, Kinose Y, Yoshimura A, Toda A, Nakatsuka E, Hashimoto K, Mabuchi S, Morishige KI, Kurachi H, Lengyel E, Kimura T. Exosomes Promote Ovarian Cancer Cell Invasion through Transfer of CD44 to Peritoneal Mesothelial Cells. Mol Cancer Res. 2017;15:78-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 305. | Menck K, Sönmezer C, Worst TS, Schulz M, Dihazi GH, Streit F, Erdmann G, Kling S, Boutros M, Binder C, Gross JC. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J Extracell Vesicles. 2017;6:1378056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 210] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 306. | Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-17452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1382] [Cited by in F6Publishing: 1491] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 307. | Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849-10859. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 497] [Cited by in F6Publishing: 563] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 308. | Matsumoto A, Takahashi Y, Nishikawa M, Sano K, Morishita M, Charoenviriyakul C, Saji H, Takakura Y. Accelerated growth of B16BL6 tumor in mice through efficient uptake of their own exosomes by B16BL6 cells. Cancer Sci. 2017;108:1803-1810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 309. | Arenz C. Small molecule inhibitors of acid sphingomyelinase. Cell Physiol Biochem. 2010;26:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 310. | Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1084] [Cited by in F6Publishing: 1213] [Article Influence: 101.1] [Reference Citation Analysis (0)] |
| 311. | Harada T, Yamamoto H, Kishida S, Kishida M, Awada C, Takao T, Kikuchi A. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci. 2017;108:42-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 312. | Catalano M, O'Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles. 2020;9:1703244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 330] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 313. | Li W, Hu Y, Jiang T, Han Y, Han G, Chen J, Li X. Rab27A regulates exosome secretion from lung adenocarcinoma cells A549: involvement of EPI64. APMIS. 2014;122:1080-1087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 314. | Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Théry C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920-4930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 399] [Cited by in F6Publishing: 465] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 315. | Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883-891. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2495] [Cited by in F6Publishing: 2721] [Article Influence: 226.8] [Reference Citation Analysis (0)] |
| 316. | Roma-Rodrigues C, Pereira F, Alves de Matos AP, Fernandes M, Baptista PV, Fernandes AR. Smuggling gold nanoparticles across cell types - A new role for exosomes in gene silencing. Nanomedicine. 2017;13:1389-1398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 317. | Shin SJ, Smith JA, Rezniczek GA, Pan S, Chen R, Brentnall TA, Wiche G, Kelly KA. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:19414-19419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 318. | Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. 2017;66:30-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 533] [Cited by in F6Publishing: 634] [Article Influence: 90.6] [Reference Citation Analysis (0)] |
| 319. | Ranganathan S, Jackson RL, Harmony JA. Effect of pantethine on the biosynthesis of cholesterol in human skin fibroblasts. Atherosclerosis. 1982;44:261-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 320. | Kavian N, Marut W, Servettaz A, Nicco C, Chéreau C, Lemaréchal H, Guilpain P, Chimini G, Galland F, Weill B, Naquet P, Batteux F. Pantethine Prevents Murine Systemic Sclerosis Through the Inhibition of Microparticle Shedding. Arthritis Rheumatol. 2015;67:1881-1890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 321. | Roseblade A, Luk F, Ung A, Bebawy M. Targeting microparticle biogenesis: a novel approach to the circumvention of cancer multidrug resistance. Curr Cancer Drug Targets. 2015;15:205-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 322. | Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5:1159-1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 382] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 323. | Sinha S, Hoshino D, Hong NH, Kirkbride KC, Grega-Larson NE, Seiki M, Tyska MJ, Weaver AM. Cortactin promotes exosome secretion by controlling branched actin dynamics. J Cell Biol. 2016;214:197-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 204] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 324. | Li B, Antonyak MA, Zhang J, Cerione RA. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;31:4740-4749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 252] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 325. | Latham SL, Chaponnier C, Dugina V, Couraud PO, Grau GE, Combes V. Cooperation between β- and γ-cytoplasmic actins in the mechanical regulation of endothelial microparticle formation. FASEB J. 2013;27:672-683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 326. | Abid Hussein MN, Böing AN, Sturk A, Hau CM, Nieuwland R. Inhibition of microparticle release triggers endothelial cell apoptosis and detachment. Thromb Haemost. 2007;98:1096-1107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 327. | Leloup L, Wells A. Calpains as potential anti-cancer targets. Expert Opin Ther Targets. 2011;15:309-323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 328. | Huang MB, Gonzalez RR, Lillard J, Bond VC. Secretion modification region-derived peptide blocks exosome release and mediates cell cycle arrest in breast cancer cells. Oncotarget. 2017;8:11302-11315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 329. | El Andaloussi S, Lakhal S, Mäger I, Wood MJ. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65:391-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 327] [Cited by in F6Publishing: 357] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 330. | Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 903] [Cited by in F6Publishing: 826] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 331. | Armstrong JPK, Stevens MM. Strategic design of extracellular vesicle drug delivery systems. Adv Drug Deliv Rev. 2018;130:12-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 332. | Yang B, Chen Y, Shi J. Exosome Biochemistry and Advanced Nanotechnology for Next-Generation Theranostic Platforms. Adv Mater. 2019;31:e1802896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 201] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 333. | Kutova OM, Guryev EL, Sokolova EA, Alzeibak R, Balalaeva IV. Targeted Delivery to Tumors: Multidirectional Strategies to Improve Treatment Efficiency. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 334. | Smyth TJ, Redzic JS, Graner MW, Anchordoquy TJ. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim Biophys Acta. 2014;1838:2954-2965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 132] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 335. | Rufino-Ramos D, Albuquerque PR, Carmona V, Perfeito R, Nobre RJ, Pereira de Almeida L. Extracellular vesicles: Novel promising delivery systems for therapy of brain diseases. J Control Release. 2017;262:247-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 336. | Das CK, Jena BC, Banerjee I, Das S, Parekh A, Bhutia SK, Mandal M. Exosome as a Novel Shuttle for Delivery of Therapeutics across Biological Barriers. Mol Pharm. 2019;16:24-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 337. | Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, Ju S, Mu J, Zhang L, Steinman L, Miller D, Zhang HG. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther. 2011;19:1769-1779. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 820] [Cited by in F6Publishing: 942] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 338. | Agrawal AK, Aqil F, Jeyabalan J, Spencer WA, Beck J, Gachuki BW, Alhakeem SS, Oben K, Munagala R, Bondada S, Gupta RC. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine. 2017;13:1627-1636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 321] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 339. | Zhang P, Zhang L, Qin Z, Hua S, Guo Z, Chu C, Lin H, Zhang Y, Li W, Zhang X, Chen X, Liu G. Genetically Engineered Liposome-like Nanovesicles as Active Targeted Transport Platform. Adv Mater. 2018;30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 340. | Li Y, Gao Y, Gong C, Wang Z, Xia Q, Gu F, Hu C, Zhang L, Guo H, Gao S. A33 antibody-functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine. 2018;14:1973-1985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 341. | Hadla M, Palazzolo S, Corona G, Caligiuri I, Canzonieri V, Toffoli G, Rizzolio F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine (Lond). 2016;11:2431-2441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 189] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 342. | Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12:655-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 615] [Cited by in F6Publishing: 870] [Article Influence: 96.7] [Reference Citation Analysis (0)] |
| 343. | Li L, Lu S, Liang X, Cao B, Wang S, Jiang J, Luo H, He S, Lang J, Zhu G. γδTDEs: An Efficient Delivery System for miR-138 with Anti-tumoral and Immunostimulatory Roles on Oral Squamous Cell Carcinoma. Mol Ther Nucleic Acids. 2019;14:101-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 344. | Lin Y, Wu J, Gu W, Huang Y, Tong Z, Huang L, Tan J. Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv Sci (Weinh). 2018;5:1700611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 280] [Cited by in F6Publishing: 310] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 345. | Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1021] [Cited by in F6Publishing: 1156] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 346. | Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782-790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 529] [Cited by in F6Publishing: 578] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 347. | Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1514] [Cited by in F6Publishing: 1562] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 348. | Aspe JR, Diaz Osterman CJ, Jutzy JM, Deshields S, Whang S, Wall NR. Enhancement of Gemcitabine sensitivity in pancreatic adenocarcinoma by novel exosome-mediated delivery of the Survivin-T34A mutant. J Extracell Vesicles. 2014;3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 349. | Jung KO, Jo H, Yu JH, Gambhir SS, Pratx G. Development and MPI tracking of novel hypoxia-targeted theranostic exosomes. Biomaterials. 2018;177:139-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 135] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 350. | Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered Exosomes for Targeted Transfer of siRNA to HER2 Positive Breast Cancer Cells. Appl Biochem Biotechnol. 2019;187:352-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 123] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 351. | Cheng Q, Shi X, Han M, Smbatyan G, Lenz HJ, Zhang Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J Am Chem Soc. 2018;140:16413-16417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 352. | Paul D, Roy A, Nandy A, Datta B, Borar P, Pal SK, Senapati D, Rakshit T. Identification of Biomarker Hyaluronan on Colon Cancer Extracellular Vesicles Using Correlative AFM and Spectroscopy. J Phys Chem Lett. 2020;11:5569-5576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 353. | Hong Y, Kim YK, Kim GB, Nam GH, Kim SA, Park Y, Yang Y, Kim IS. Degradation of tumour stromal hyaluronan by small extracellular vesicle-PH20 stimulates CD103+ dendritic cells and in combination with PD-L1 blockade boosts anti-tumour immunity. J Extracell Vesicles. 2019;8:1670893. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 354. | Piffoux M, Nicolás-Boluda A, Mulens-Arias V, Richard S, Rahmi G, Gazeau F, Wilhelm C, Silva AKA. Extracellular vesicles for personalized medicine: The input of physically triggered production, loading and theranostic properties. Adv Drug Deliv Rev. 2019;138:247-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |