Copyright
©The Author(s) 2021.
World J Gastrointest Oncol. Jul 15, 2021; 13(7): 673-683
Published online Jul 15, 2021. doi: 10.4251/wjgo.v13.i7.673
Published online Jul 15, 2021. doi: 10.4251/wjgo.v13.i7.673
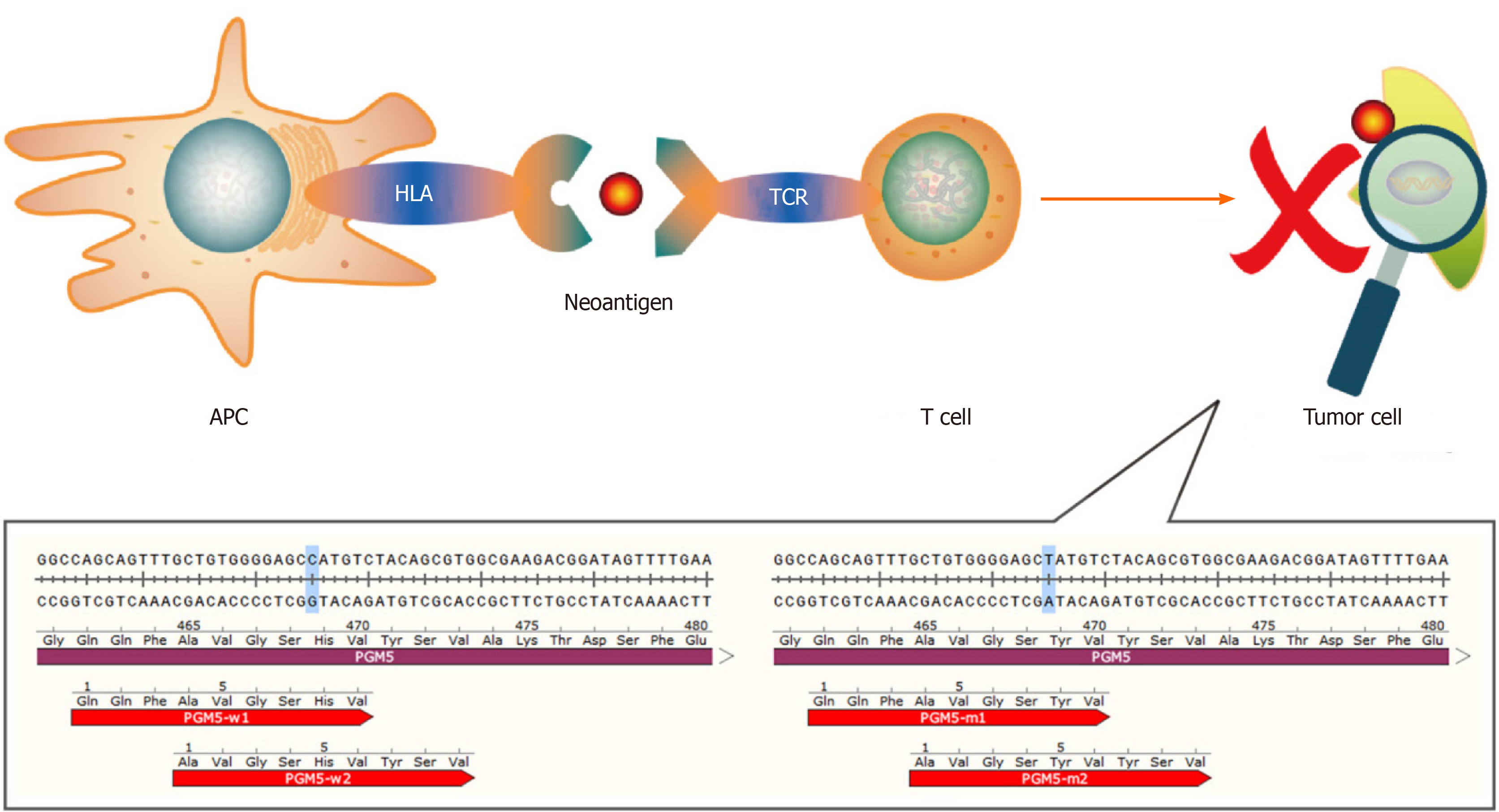
Figure 1 The human PGM5 gene, as an example.
Antigen-presenting cells present neoantigens to T cells through human leukocyte antigen molecules, and the T cell receptor recognizes neoantigens. This recognition activates T cells to eliminate neoantigen-specific tumor cells. The bottom part of the figure shows mutations of the PGM5 gene that produce two neoantigens. APC: Antigen-presenting cell; HLA: Human leukocyte antigen; TCR: T cell receptor.
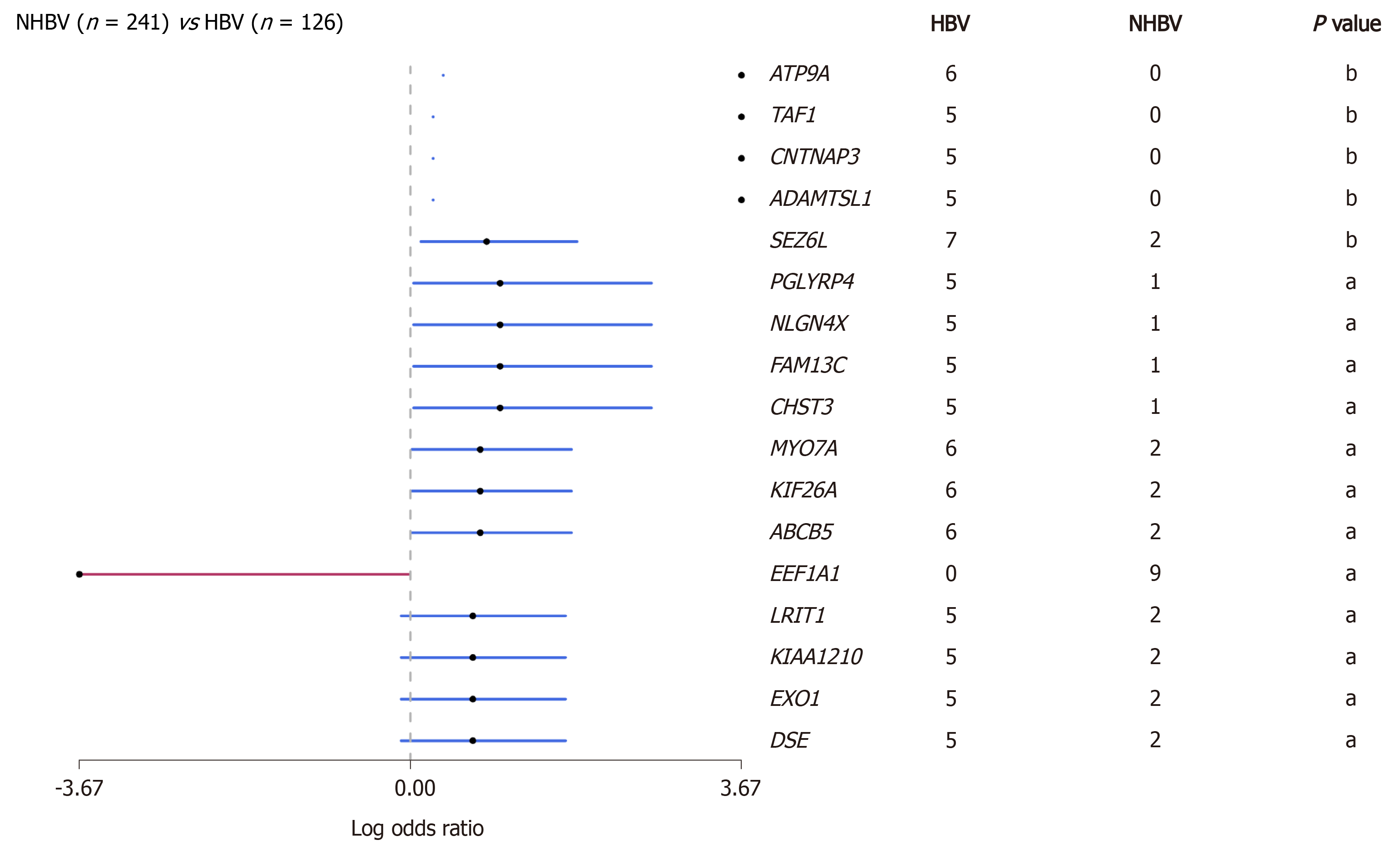
Figure 2 Comparison of gene mutations between hepatitis B virus-related hepatocellular carcinoma and non-hepatitis B virus-related hepatocellular carcinoma in data from the The Cancer Genome Atlas database (aP < 0.
05; bP < 0.01). The red line represents genes in non-hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) cohort, and the blue line represents genes in HBV-related HCC cohort. HBV: Hepatitis B virus.
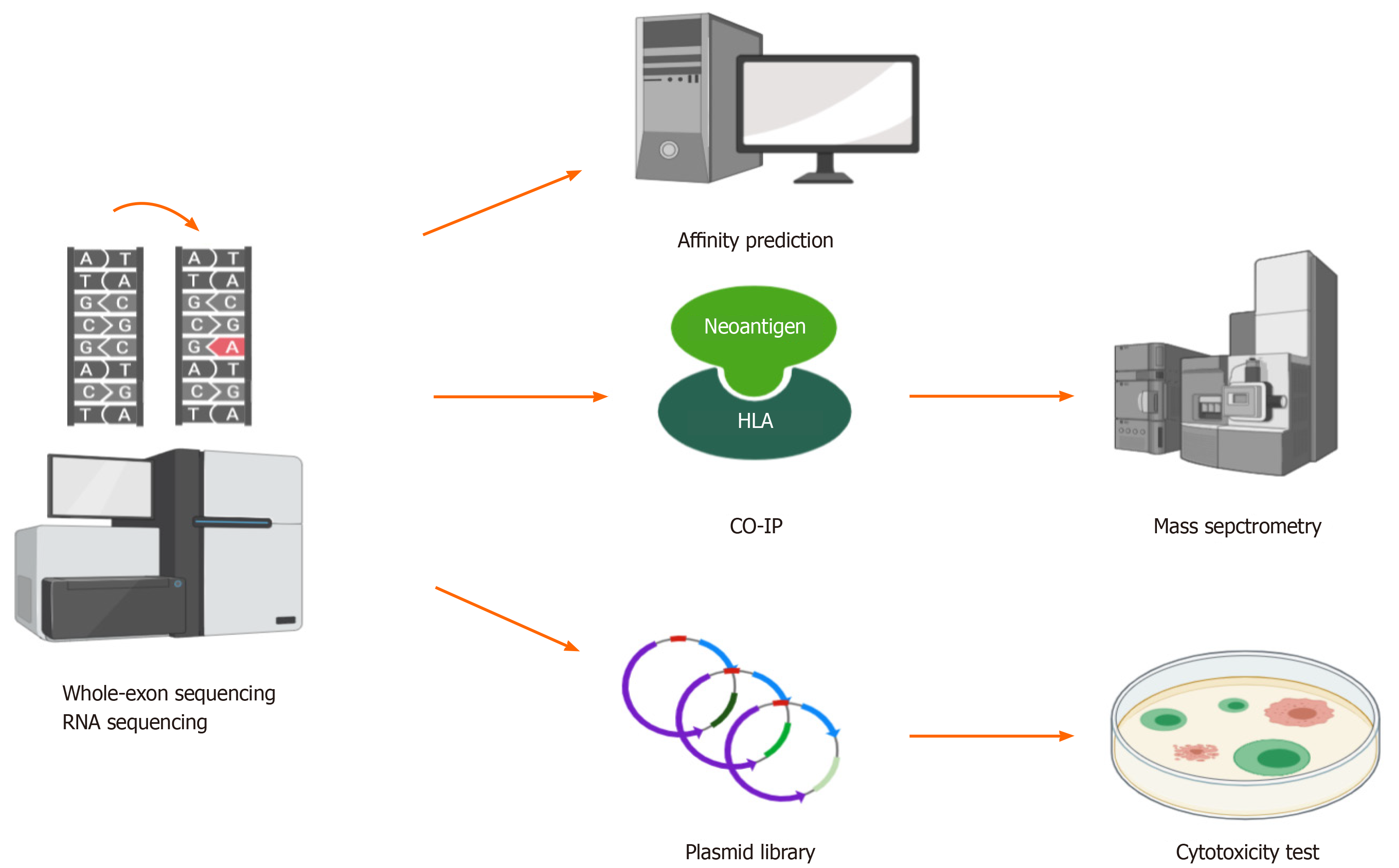
Figure 3 Whole-exon sequencing and RNA sequencing detect mutations in tumors, and then, different methods can be used to identify hepatocellular carcinoma neoantigens.
(1) Human leukocyte antigen affinity prediction; (2) Coimmunoprecipitation followed by mass spectrometry; and (3) Construction of a plasmid library based on neoantigens followed by cytotoxicity tests in vitro. HLA: Human leukocyte antigen; CO-IP: Coimmunoprecipitation.
- Citation: Chen P, Fang QX, Chen DB, Chen HS. Neoantigen vaccine: An emerging immunotherapy for hepatocellular carcinoma. World J Gastrointest Oncol 2021; 13(7): 673-683
- URL: https://www.wjgnet.com/1948-5204/full/v13/i7/673.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i7.673