Peer-review started: July 9, 2020
First decision: October 6, 2020
Revised: December 10, 2020
Accepted: December 26, 2020
Article in press: December 26, 2020
Published online: January 27, 2021
Processing time: 196 Days and 2.7 Hours
Autophagy is the liver cell energy recycling system regulating a variety of homeostatic mechanisms. Damaged organelles, lipids and proteins are degraded in the lysosomes and their elements are re-used by the cell. Investigations on autophagy have led to the award of two Nobel Prizes and a health of important reports. In this review we describe the fundamental functions of autophagy in the liver including new data on the regulation of autophagy. Moreover we emphasize the fact that autophagy acts like a two edge sword in many occasions with the most prominent paradigm being its involvement in the initiation and progress of hepatocellular carcinoma. We also focused to the implication of autophagy and its specialized forms of lipophagy and mitophagy in the pathogenesis of various liver diseases. We analyzed autophagy not only in well studied diseases, like alcoholic and nonalcoholic fatty liver and liver fibrosis but also in viral hepatitis, biliary diseases, autoimmune hepatitis and rare diseases including inherited metabolic diseases and also acetaminophene hepatotoxicity. We also stressed the different consequences that activation or impairment of autophagy may have in hepatocytes as opposed to Kupffer cells, sinusoidal endothelial cells or hepatic stellate cells. Finally, we analyzed the limited clinical data compared to the extensive experimental evidence and the possible future therapeutic interventions based on autophagy manipulation.
Core Tip: Extensive investigation of autophagy is mostly based on experimental data. However there is now enough evidence to support the notion that autophagy is not only the waste recycling mechanism of the hepatocyte, but is strongly involved in the pathogenesis of almost all liver diseases. It can be either a defensive mechanism against various insults or a detrimental machinery aggravating the underlying disease. Modulation of autophagy has different consequences in the hepatocyte than in the liver macrophages, the sinusoidal endothelium or the hepatic stellate cells. There is also an opportunity for future treatment applications of autophagy manipulation.
- Citation: Kouroumalis E, Voumvouraki A, Augoustaki A, Samonakis DN. Autophagy in liver diseases. World J Hepatol 2021; 13(1): 6-65
- URL: https://www.wjgnet.com/1948-5182/full/v13/i1/6.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i1.6
Autophagy (from the Greek self-eating) is a process crucial for cell survival[1,2]. Autophagy is a lysosomal degradation pathway that controls the disposition of intracellular waste including damaged organelles or invading pathogens. It can be characterized as the recycling energy system of the cell.
Under basal conditions autophagy degrades 1.5% of total hepatic protein per hour but in starvation, protein degradation increases to 4.5% of liver protein per hour[3]. When rodents are starved for 48 h, autophagy degrades up to 40% of liver protein[4].
Although It is accepted that the term “autophagy” was introduced in 1963 by the Belgian researcher Christian René de Duve, in fact the term autophagy was used almost a century earlier by Anselmier in a French journal[5].
However the modern era of autophagy started with the pioneer work of de Duve and Novicoff in the 1950s when acid phosphatase positive lysosomes were described in the rat liver[6-9] and the term lysosome was used for the first time[10].Later de Duve introduced the term autophagosome and Arstila and Trump proved that the autophagosomes originate from the endoplasmic reticulum (ER)[11]. The next important progress came when Takeshige et al[12] identified approximately fifteen Autophagy related genes (Atgs) involved in Saccharomyces cerevisiae autophagy[12-14]. Today, more than 40 Atgs in various animal and human cells have been identified and unified[15-17]. The importance of autophagy was recognized by the award of two Nobel Prizes for Physiology or Medicine, the first to Cristian De Duve in 1974 and the second to Yoshinori Ohsumi in 2016[18,19]. Landmarks of autophagy were recently described[20]. During the period 2008-2018 more than 33000 papers related to autophagy were published[21,22].
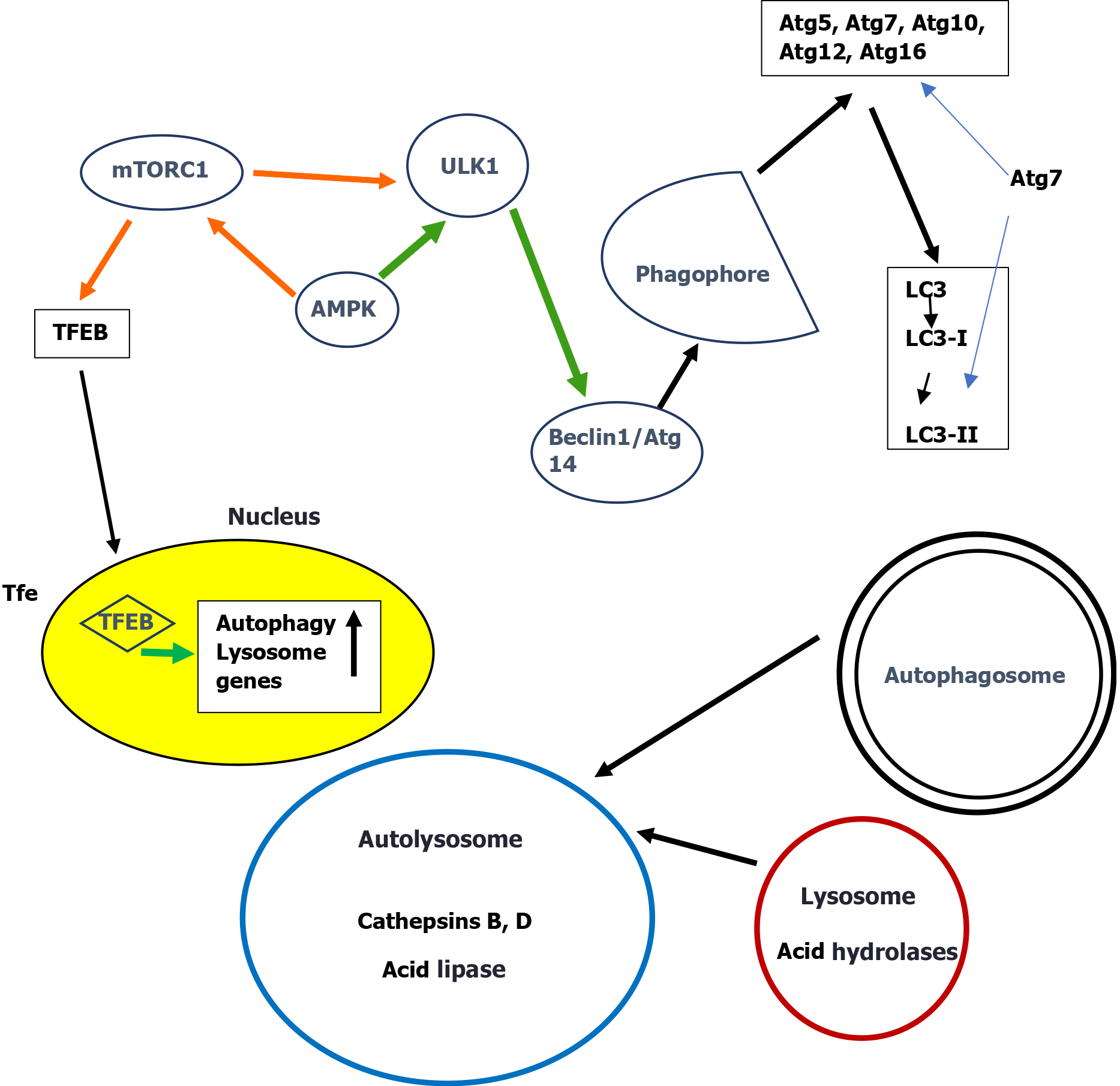
Autophagy has certain discrete stages including induction, phagophore formation, autophagosome formation, autolysosome formation and degradation[23-25]. Atg molecules are involved in various complexes essential for autophagy induction and autophagosome formation[26]. Initiation starts with activation of the unc-51-like kinase 1 complex (ULK1, Atg1 in yeast) followed by beclin 1 (Atg6 in yeast) and a subsequent cascade of Atg proteins leading to autophagosome formation where LC3 (Atg8 in yeast) is implicated[27]. LC3 is further processed to form initially LC3-I and then LC3-II[28]. Once the autophagosome is formed, a blockage of autophagic flux at late steps will downregulate the clearance of autophagosomes. A blockage of autophagic flux finally results in autophagy dependent cell death[29]. Detailed descriptions of the complex molecular steps of each stage of autophagy were recently published[20,28,30].
A commonly used marker for estimating autophagosome formation is the fusion protein green fluorescent protein-LC3 (GFP-LC3)[31]. Of the three members LC3A, LC3B, and LC3C of the human LC3 gene family, LC3B and LC3-II are mostly used for autophagy assays[32-34]. Autophagic flux into the lysosomes is estimated by measuring p62/SQSTM1 degradation. p62/SQSTM1 is a protein complex that binds to LC3 and is efficiently degraded by autophagy[35]. The total cellular level of p62/SQSTM1 inversely correlates with autophagic activity. Thus in autophagy-deficient cells, p62/SQSTM1 levels are increased after starvation in contrast to cells with normal autophagy[36].
It should be stressed that he level of LC3 is related to the induction of autophagy but might not reflect the final stages of autophagy and should not be used as a general marker of autophagy[34-36]. Further progress of autophagy is detected by a low level of p62 since p62 degradation depends on the function of the autophagosome-lysosome fusion[37]. Therefore an increase of both LC3 and p62 indicates formation of autophagosomes without lysosomal degradation[38].
As mentioned before, a major breakthrough in autophagy was the identification of Atgs. Evidence for the importance of autophagy in liver homeostasis was provided by the generation of of Atgs-knockout mice models[39]. Livers of mice with deletion of the autophagy gene Atg7 were markedly enlarged, up to 30% of the body weight of the animal and hepatocytes were characterized by structural alterations of mitochondria and peroxisomes and aggregation of ubiquitinated proteins. These aggregates disappeared when the ATg7- knockout mouse was bred to a mouse null for SQSTM1/p62indicating that SQSTM1 is important to direct damaged cytosolic proteins into the autophagic pathway[40,41].
To date, three major types of autophagy, namely, macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA), have been described[22,42,43].
Macroautophagy is the classical pathway that engulfs the cytosolic components targeted for lysosomal degradation. Initiation of autophagy is controlled by two metabolic sensors the mammalian target of rapamycin complex 1 (mTORC1) and the AMP-activated protein kinase (AMPK). mTORC1 negatively regulates autophagy by direct phosphorylation of ULK1 thus inhibiting ULK1. AMPK suppresses mTORC1 activity by phosphorylation of tuberous sclerosis 2 and raptor, two essential regulators of mTORC1[44,45]. Recently it was reported that the final step in this activation process of mTOR is dependent on Rheb, a small GTPase that binds to mTOR and allosterically activates its kinase activity[46]. The long-term regulation of autophagy is carried out by transcription factor EB (TFEB)[47], the main regulator of lysosomal biogenesis and autophagy. Under nutrient-rich conditions, mTORC1 phosphorylates TFEB and retains TFEB in the cytosol[48-50]. Nutrient deprivation on the other hand leads to mTORC1 inhibition, dephosphorylation of TFEB and its translocation to the nucleus to initiate the rapid transcription of autophagy genes[51,52]. All subsequent series of complex events leading to the final degradation in lysosomes have elegantly been described[2,24,53].
A simplified scheme of macroautophagy is presented in Figure 1.
Microautophagy is the least studied type of autophagy where compounds or membranous vesicles are directly taken up by lysosomes[54]. Microautophagy is important during amino acid starvation[55,56] and possibly three different types can be recognized[57].
Chaperone Mediated Autophagy (CMA) is a selective engulfment process of substrates containing the pentapeptide “Lys-Phe-Glu-Arg-Gln” (KFERQ) motifs. They are recognized by, the cytosolic chaperone heat-shock cognate protein of 70 kDa (HSC70), and transported into the lysosomes through the lysosomal membrane protein 2A (LAMP2A)[58,59]. CMA is induced by DNA damage, hypoxia and oxidative stress, among others[60-65].
Today macroautophagy is also divided into non selective autophagy and selective macroautophagy targeting special organelles or specific compounds for degradation[43,66,67]. Thus new names have appeared according to the coumpounds involved: Ribophagy (ribosomes)[68], pexophagy (peroxisomes)[69], ferritinophagy (iron-based compounds)[70] and most importantly reticulophagy (ER)[71] lipophagy (lipids)[72] and mitophagy (mitochondria)[73]. The last two are practically involved in every form of fatty liver.
Reticulophagy: Multiple receptors directly interact with LC3 and form autophagosomesduring reticulophagy, a very important form of macroautophagy thatpreserves the size and function of the ER in different conditions like starvation, non-alcoholic fatty liver disease (NAFLD), viral infections and fibrosis[74-79].
Lipophagy: Lipophagy is implicated in lipid homeostasis and metabolism in liver diseases. It is usually down-regulated in steatosis of either alcoholic or non-alcoholic liver disease[80-84], but it is up-regulated when fibrosis, cirrhosis or hepatocellular carcinoma are evolving[85-87]. Comprehensive reviews of lipophagy in liver disease were recently presented[88-91].
Mitophagy: The first step of mitophagy in mammals requires the induction of canonic Atg-dependentautophagy with either mTOR suppression induced by mitochondrial generated reactive oxygen species (ROS), or AMPK activation induced by adenosine triphosphate (ATP) depletion. The second step is the priming of the mitochondria involving molecular modifications leading to their recognition by the autophagosomes[92,93]. Even in the healthy liver, worn out mitochondria with a half-life of 10 to 25 d are removed by mitophagy[94,95]. Elimination of aged or damaged mitochondria protect cells from release of pro-apoptotic proteins, generation of toxic ROS and non proper hydrolysis of ATP[96-99]. When oxidative stress appears, autophagy rapidly acts to remove oxidized proteins or damaged mitochondria that generate more ROS. Recent data show that in autophagy deficiency there is acummulation of ROS and p62 probably mediated by the loss ofFOXO1/3. It has been reported that the p62-FOXO1/3 axis is the molecular basis for the reduction of antioxidant defense in autophagy deficiency[100]. Three different types of mitophagy have been described based in the different molecular pathways involved[101,102]. An extensive review of molecular mechanisms of mitophagy in liver diseases has been recently published[103].
New players in liver autophagy: It is clear today that apart from the known pathways regulating liver autophagy, there are additional mechanisms involved. The most important are the long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and exosomes. Many recent studies have presented strong evidence that ncRNAs influence autophagy by regulating various autophagy pathways[104-110]. Equally, miRNAs regulate autophagy influencing the core autophagy pathways[111].
Evidence from experimental animals with liver specific deletions of Atgs has demonstrated the role of High mobility group box 1 (HMGB1)[112] and Yes-associated protein (YAP)[113] in the pathological changes induced by autophagy. Nuclear receptors were also reported to control autophagy. Activation of the farnesoid X receptor (FXR), occurs during feeding and suppresses Atgs expression. On the other hand during starvation, fasting-activated nuclear receptors, the peroxisome proliferator-activated receptor alpha (PPAR), and the cAMP response element-binding protein (CREB), induce expression of Atgs and therefore increase autophagy[114-116].
An association of autophagy with the formation and function of exosomes has also been described. Exosomes are extracellular vesicles originating from late endosomes, which do not fuse with lysosomes but are released extracellularly by exocytosis. Exosomes can either activate autophagy pathways or transfer extracellular vesicles to the lysosomes[117].The interplay between autophagy and exosome biogenesis has been recently described[118].
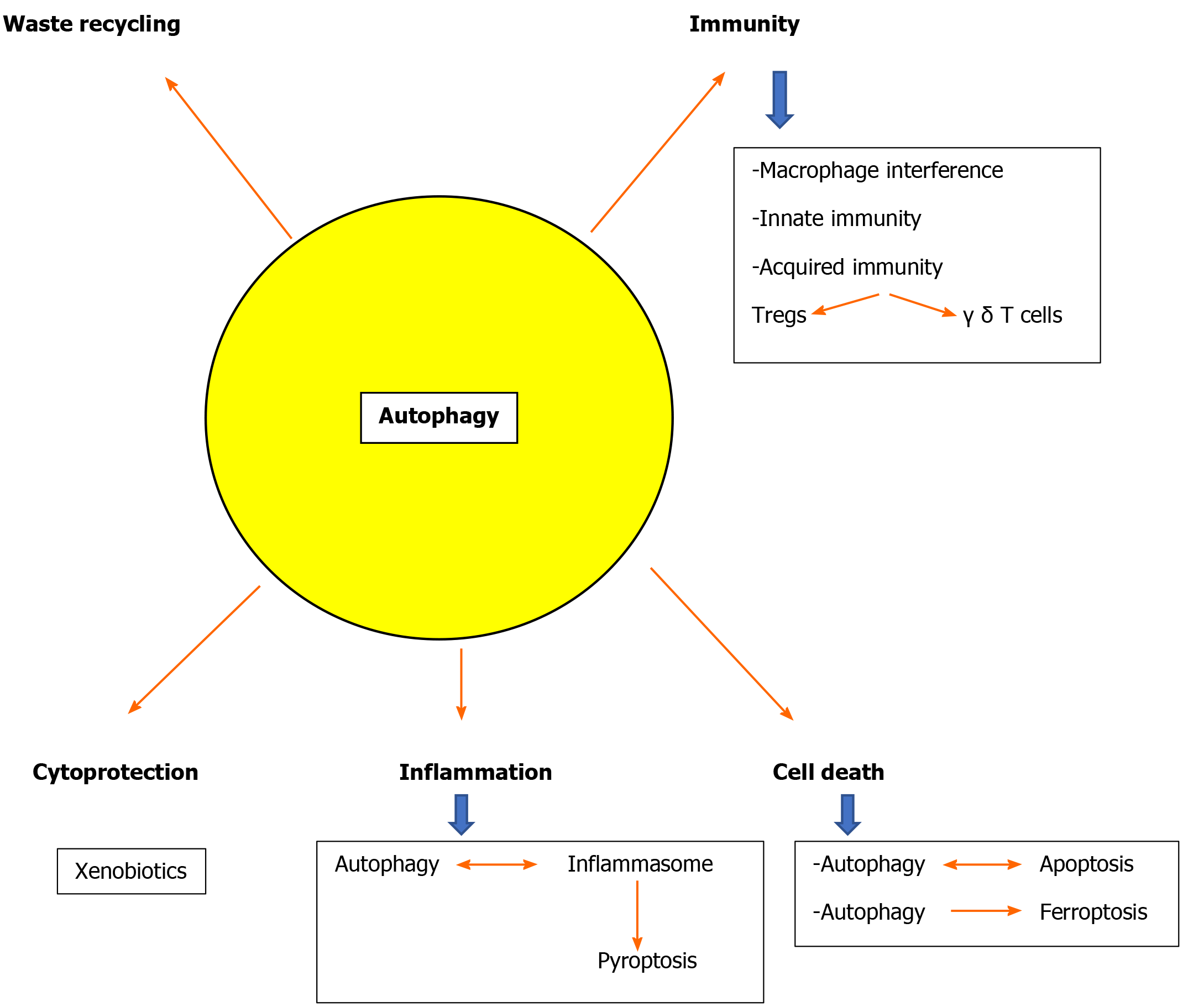
Most researchers have studied either the early or the late stages of autophagy. However equally important is the final stage, namely the lysosome reformation (ALR), leading to regeneration of functional lysosomes from autolysosomes. A series of proteins including clathrin, the motor protein KIF5B, and dynamin 2 are sequentially involved up to the maturation of functional lysosomes. Early lysosomes are pH-neutral but eventually they gain acidity and luminal proteins[119-122]. Accumulating evidence suggests that most, if not all, components of the molecular machinery for autophagy also mediate autophagy-independent functions. Autophagy is involved in various cell functions like endocytosis, phagocytosis, DNA repair, centrosome function, cell proliferation, cell death and immunological response including memory. Details were recently reported[123].
Autophagy and immunity: The implication of autophagy with the immune system has been investigated in the last few years[124-131]. Non-canonical forms of macroautophagy were described, resulting in the formation of autophagosomes that fuse with the lysosomes[132]. Only a subset of the Atgs machinery is used. Among these, LC3-associated phagocytosis (LAP) has been extensively studied because of its implication in immune regulation. LAP recruits LC3-II to the phagosomal membrane[133-135] and is taken up by macrophages through innate immune receptors such as Toll-like receptors. In contrast to autophagy the LAPasome is a single membrane vacuole. In contrast to autophagy, ULK1 is not required for LAP[133]. Chaperone-mediated autophagy has also attracted attention because of its central role in antigen presentation and aging[136,137]. Autophagy is also implicated in the function of innate immunity interfering with macrophage autophagy. There is interplay between autophagy and innate immunity as interferon (IFN)-γpromotes autophagy in macrophages[138]. Mice fed with high fat diet had impaired autophagy in bone marrow-derived macrophages and peritoneal macrophages[139]. Mice with Atg5 deficient macrophages, developed hepatic inflammation when stimulated with lipopolysaccharide (LPS) after a high fat diet feeding. Acquired immunity is primarily a defense function against specific pathogens and is brought about by the different subsets of T cells and B cells. Interestingly there is evidence that high autophagic activity maintains the differentiation and function of important T-cell subsets such as regulatory T (Treg)-cells[140] and γ δ T-cells[141].
Autophagy and cell death: It has been proven that autophagy can be either a protective mechanism or a contributor to cellular death in certain instances[142-144]. Autophagy is involved in cellular death mostly by its effects on apoptosis. Autophagy is connected to apoptosis and these two cellular destructive phenomena are affecting each other[145-148]. This is particularly important in hepatic cell death[149].
Generally autophagy blocks the induction of caspase-dependent apoptosis, and apoptosis-associated caspase activation stops the autophagic process. Yet, in special cases, autophagy may induce apoptosis or necrosis, and autophagy has been shown to degrade the cytoplasm, leading to ‘autophagic cell death’[150-152].
Autophagy is also implicated in caspase-independent cell death, leading to necrosis and necroptosis[153]. Induction of apoptosis eliminates cells damaged through the action of the tumor suppressor gene p53[154]. Apoptosis is counteracted, among others, by the mTOR/AKT pathway also involved in autophagy. The balance between p53 and AKT/mTOR is crucial for the fate of injured cells[155,156]. In addition, autophagy induces a particular mechanism of cell death named ferroptosis. It was initially reported as a specific iron-dependent form of malignant cell death. It soon became clear that ferroptosis is a more general form of cell death[157,158]. Many proteins implicated in autophagy (like Atgs and BECN1) were also involved in ferroptosis. Moreover activators of ferroptosis, like erastin, induced autophagosome accumulation and activation of autophagy led to ferroptotic cell death possibly by the turnover of ferritin through ferritinophagy[159-161].
A recent study has shown that ferroptosis is also interconnected with lipophagy. Lipids released during lipophagy and subsequent peroxidized increase ferroptosis. Therefore it might be that ferroptosis is a mechanism of cellular death in NAFLD[162].
Autophagy and inflammation: Autophagy is also closely associated with the inflammatory response in the liver. Inflammasome and autophagy regulate each other by the same inhibitory mechanisms which however are controlled by different input pathways. The NLRP3 inflammasome activation, usually through the stimulation by pathogen- and/or danger-associated molecular patterns[163,164], induces procaspase-1 activation which promotes interleukin interleukin (IL)-1β and IL-18 production leading to pyroptotic cell death. These events are counteracted by caspase-1-mediated activation of autophagy. In addition autophagy reduces inflammasome activation degrading the inflammasomes in the autophagosomes but also eliminating damaged cytoplasmic organelles that otherwise would produce DAMPS increasing activation of inflammasomes[165,166].
On the other hand, the negative correlation between inflammasomes and autophagy[167-169] leads to an increased production of the pro-inflammatory IL-1β[170] when autophagy is decreased[128].However, the relationship between NLRP3 and autophagy has not been fully clarified, and recent studies have reported that nuclear factor-κappa beta (NF-κB) activation can modulate the NLRP3 and autophagy towards the same direction[171].
In view of the above is not surprising that many reviews on autophagy use the term “double-edged sword” stressing the fact that autophagy may have opposite effects on the same biological phenomenon[172]. Prominent general paradigms are cancer[173,174] and viral infections[175].
Another characterization pertinent to the liver is that autophagy behaves like Jekyll and Hyde depending on the cells involved. In hepatocytes, macroautophagy [in NAFLD and alcoholic liver disease (ALD)] and CMA (in NAFLD) is protective. It reduces fat accumulation and oxidative stress, it removes damaged mitochondria and favors regeneration. In macrophages, macroautophagy inhibits liver inflammation and fibrosis but it enhances fibrosis activated stellate cells. It is protective in early phases of hepatocellular carcinoma, but may be detrimental in late phases[176,177].
Autophagy in hepatocytes but also in the non-parenchymal sinusoidal cells of the liver is a key for liver physiology[178,179] and defects of autophagy are implicated in the pathophysiology of most liver diseases[180]. Both common diseases like alcoholic and non-alcoholic fatty liver or viral hepatitis and rare entities like Wilson’s disease and a1 antitrypsin deficiency are related to autophagy defects[30,41,57,181-184]. Defective autophagy also leads to accumulation of detrimental hepatocyte byproducts due to the fact that hepatocytes have a long half life of 6-12 mo [143]. Moreover, the liver is responsible for handling of a large number of xenobiotics and autophagy is a cytoprotective mechanism[99,185] (Figure 2).
NAFLD is the commonest liver disease worldwide. Recently it was suggested that it should be renamed as metabolic dysfunction-associated fatty liver disease (MAFLD)[186,187]. Pathological lesions in the liver vary from simple steatosis to non-alcoholic steatohepatitis (NASH) and cirrhosis. Current pathogenesis of NASH is mainly focused on the effects of insulin resistance and lipotoxicity in hepatocytes[188]. The abnormalities reported in Kupffer cells, stellate cells and endothelial cells are regarded as secondary events[189,190].
Obesity and insulin resistance are well documented risk factors for NAFLD development. Defects in liver autophagy have been established as fundamental abnormalities in both conditions.
In the hepatocyte, lipids are catabolized by two major pathways. The first involves cytoplasmic neutral lipases and the second is lipophagy and acid lipases and hydrolases of the lysosomes. The end result is the production of free fatty acids that are further broken down by βI-oxidase in the mitochondria[191].
Lipid droplets have a core of lipids enwrapped in a phospholipid layer characterized by proteins called perilipins directing them to the autophagosome[72]. A crucial protein mediating lipolysis and autophagy is the adipose triglyceride lipase (ATGL). Cytoplasmic lipolysis and lipophagy are interconnected. The degradation of perilipins by autophagy facilitates actions of ATGL which in turn induces autophagy via sirtuin1 deacetylation of certain Atgs and activation of the transcription factors FoxO1 and FoxO3 thus promoting autophagy[192-194].
Lipophagy can prevent lipid accumulation in hepatocytes, while the inhibition of lipophagy promotes lipid droplets (LDs) accumulation, resulting in hepatocellular steatosis[195].
Characteristic changes of the metabolic syndrome like obesity, hyperglycemia, and dyslipidemia have been shown to exert a negative effect on autophagy because the regulatory control of forkhead box O1 (FoxO1) on the expression of Atg genes is lost leading to autophagy malfunction[196]. Macroautophagy and CMA are also down-regulated by increased intracellulal lipids due to either interference with the lysosomal stability of the CMA receptor or to the reduction of the ability of autophagosomes to fuse with lysosomes leading to the reduction of macroautophagic flux[196-198].
The severity of steatosis is related to the expression of three proteins, the damage regulated autophagy modulator (DRAM), BAX and p53. In mice livers, p53 expression increased in mild and severe steatosis. A DRAM expression increase was observed in mild hepatosteatosis, whereas high BAX expression was identified in severe hepatosteatosis[199].
A clinical study has confirmed the link between induction of autophagy and liver steatosis[200]. Autophagy-related genes (Atg5, LC3A, and LC3B) were overexpressed in obese patients compared with non obese patients.
Experimental evidence also suggests that defective autophagy is crucial in the development of obesity, oxidative stress, and the metabolic syndrome[201-203].
Insulin is intimately involved in autophagy regulation as the mTOR inhibitor of the FoxO and TFEB controllers of the transcription of autophagic genes is insulin-inducible[204]. Overactivation of mTOR in turn leads to insulin resistance[205,206]. Several mechanisms might explain this defect in obesity. Obesity increases calpain-2 by a still unknown signal pathway. Calpain is a protease that degrades Atg7 and modulates autophagy[201]. Autophagosome-lysosome fusion is also defective in livers of obese mice due to alterations of the lipids in cellular membranes induced by the high-fat diet[198]. A defective liver autophagy and the associated decrease of lysosomal degradation contribute to an additional increase in the ER stress which leads to insulin resistance and a vicious circle is completed[201,207,208]. Hyper-insulinemia decreases liver autophagy and reduced hepatic autophagy aggravates ER stress and insulin resistance.
An additional mechanism is a defect in acidification of lysosomes. Impaired substrate degradation in autolysosomes has also been reported for obese ob/ob mice. Activities of lysomal cathepsins were implicated in obesity. Cathepsin L was decreased in obese adipose tissue, while Cathepsin B was significantly elevated. Interestingly in obese adipose tissue inflammasomes were activated and further upregulation of cathepsin B resulted in additional activation of inflammasomes[209-212].
A study of the expression of 322 lysosomal/autophagic genes was recently reported in adipose tissue of lean and obese patients. Among 35 significantly expressed genes, 34 were upregulated. In isolated murine cells, tumor necrosis factor alpha (TNFα) stimulation resulted in upregulation of lysosomal/autophagic genes accompanied by upregulation of the autophagy associated SQSTM1/p62 receptor leading to increased degradation of perilipin 1. It seems that local inflammatory cytokines may impair lipid storage via autophagy induction[213].
An extensive review of lysosomal enzyme abnormalities in both adipose and liver tissue was recently published[214]. A recent report suggests an additional mechanism contributing to obesity-associated abnormalities. Obesity increases lysosomal iNOS and NO production leading to exacerbation of lysosomal nitrosative stress, impairment of lysosomal function, defective autophagy and insulin resistance[215].
There is also evidence that mitophagy is negatively regulated by liver insulin resistance. Mitophagy can promote mitochondrial fatty acid oxidation to inhibit hepatic fatty acid accumulation and improve hepatic insulin resistance. Fundc1 is a recently characterized mitophagy receptor and mice lacking this receptor develop severe obesity and insulin resistance when maintained in a high-fat diet[216,217].
However, when autophagy is defective an alternative mechanism protects the liver from steatosis. An induction of fibroblast growth factor 21 (FGF21) was reported in mice with subsequent amelioration of insulin resistance and decreased diet-induced obesity[218,219]. This has been corroborated in a clinical study of overweight NAFLD patients, where increased FGF21 levels were correlated with steatosis grade, fibrosis and lobular inflammation. NASH patients had the highest levels[220]. An analogue of FGF21 has been tested in experimental animals and obese diabetic patients with promising results[221-223]. Nevertheless, the control of adipose tissue biology is very complex and is elegantly described in a recent publication[224].
Not surprisingly autophagy is strongly associated with NAFLD pathogenesis[179]. Diet-induced NAFLD in mice blocks hepatic autophagy and leads to oxidative stress and mitochondrial dysfunction[225], also reducing thyroid hormone-induced mitophagy[226]. The potential molecular pathways and possible therapeutic implications of thyroid hormones in NAFLD have been recently reviewed[227].
Mitophagy abnormalities are strongly implicated in NAFLD[228-230]. In particular an impairment of mitophagy seems to activate the NLRP3 inflammasome favoring the progression of NAFLD to NASH[38]. Accordingly, recent evidence indicates that restoration of mitophagy may improve NAFLD[231-234].
In addition to mitophagy, reticulophagy is also implicated in NAFLD. An extensive reticulophagic response is evident in hepatocytes after induction of NAFLD by oleic acid[228,235]. It is suggested that reticulophagy and mitophagy are independent, events involved in NAFLD progression[228].
Impaired lipophagy and lipotoxicity are also strongly involved in NAFLD[72,192,236,237]. Lipid accumulation in hepatocytes blocks autophagic flux and impaired autophagic flux favors the progress of NAFLD[30].
This impaired flux and the subsequent ER stress can be improved by inhibition of the sterol regulatory element-binding protein 2 (SREBP-2) whose activation promotes accumulation of cholesterol in NAFLD. This improvement is associated with upregulation of autophagy genes[238].
Intracellular lipid trafficking is also regulated by store operated calcium entry and enhanced lipophagy is observed in cells defective in this system[239]. Moreover, the detrimental effects of diets rich in saturated FFA were increased bysirtuin-3, which enhanced lipotoxicity, reducing the autophagic flux[240]. The effect of lipophagy in liver steatosis is further supported by experimental evidence that various chemicals are involved in steatosis by interfering with autophagy. Caffeine reduces lipid content and stimulates beta-oxidation in hepatocytes through autophagy in mammalian liver cells in NAFLD[17]. In essence caffeine protects against fatty liver through the co-ordination of the induced lipophagy and mitochondrial β-oxidation[241,242]. Epidemiologic studies demonstrated that coffee consumption reduced the development of fatty liver, fibrosis, and hepatocellular carcinoma in NAFLD patients[243,244] supporting thus the experimental evidence.
Methionine is a well known inactivator of autophagy and lipophagy. The correlation between lipophagy and methionine in the liver from patients with liver steatosis has been studied. Increased levels of methionine inhibit autophagic catabolism of lipids and contribute to liver steatosis in NAFLD[83]. Mice fed with a methionine/choline deficient diet developed steatosis, inflammation, fibrosis and ER stress associated with mitochondrial dysfunction. The administration of the autophagy enhancer rapamycin ameliorated these lesions while chloroquine, a well established autophagy inhibitor, aggravated the liver injury[245]. Resveratrol, another autophagy activator, also attenuated liver lesions induced by a similar diet[246,247]. Consistent with these findings is a recent report that a traditional Chinese herb increased autophagy and considerably improved steatohepatitis induced by methionine/choline deficient diet in rats[248].
Other diet-supplied molecules affect autophagy and are possibly beneficial in NAFLD including the purple sweet potato color[249]. Likewise, the caffeic acid of vegetables has been reported to ameliorate hepatic steatosis[250] while curcumin, an antioxidant polyphenol of Curcuma longa, has been shown to inhibit apoptosis and induce autophagy with a potential protective effect on hepatocellular carcinoma[251].
A finding that might be useful in future treatment of NAFLD was recently reported. Celecoxib, a COX-2 inhibitor, attenuated steatosis and restored autophagic flux in cells treated with palmitate and rats fed a high fat diet[252].
Other lipids like the sphingolipid ceramide may be implicated in NAFLD as it is increased in Atg7 knockout mouse liver in parallel with the impaired autophagy[253]. Autophagy increased when sphingolipid de novo synthesis was upregulated, indicating that lipid degradation was activated to prevent excessive sphingolipid accumulation.
Interestingly, autophagic activity seems to be upregulated when the renin angiotensin system is overexpressed. The underlying mechanisms and its role in NAFLD have yet to be clarified as there are many controversial issues to be solved[254]. Overall there is extensive evidence that inhibition of lipophagy is detrimental for the liver in NAFLD[198,222,238,255].
Summarizing the above studies, a therapeutic approach against NAFLD would be the activation of lipophagy[90]. However, it is noteworthy that there is one study indicating the opposite, as suppression of autophagy through inhibition of c-Jun N-terminal Kinase (JNK) ameliorates insulin resistance in a rat NAFLD model[256].
Extensive reviews on the mechanisms of autophagy deregulation in NAFLD were recently published[183,257,258]. Not only impaired macroautophagy but also reduced liver chaperon mediated autophagy (CMA) favors steatosis due to failure in the timely removal of perilipins[259,260] and therefore an increase in lipogenic enzymes. When oxidative stress is increased in the liver, an upregulation of CMA occurs to selectively remove damaged proteins[62]. Loss of CMA leads to impairment of proteostasis and accumulation of oxidized protein aggregates perpetuating thus chronic oxidative stress[261].
Involvement of autophagy in the progression of NAFLD to NASH has not yet been clarified and molecular mechanisms are not fully understood.
One of the histological characteristics of NASH used in diagnosis and scoring systems is the formation of Mallory-Denk bodies (MDB)[262-264]. There is experimental evidence that inhibition of autophagy and accumulation of p62 is related to their formation while autophagy activation with rapamycin leads to their resolution[265]. Further support of the involvement of autophagy in NAFLD evolution to NASH was reported in a clinical and experimental study where a decrease of autophagic flux in parallel with an increase in ER stress was demonstrated both in the livers from NAFLD patients and mice models of NAFLD, and in lipid-overloaded human hepatocytes[266]. However tests for measurements of autophagic flux used in this paper are not full-proof as they can be influenced by autophagy independent factors. Therefore these findings should be corroborated in a different set up.
Patients with NASH and murine models of steatotic inflammation had reduced expression of Atg7 and TFEB while the autophagy inhibitor rubicon was increased[139,177,255].
In contrast, steatosis and liver injury were improved in parallel with restoration of autophagy and reduction of ER stress in mice with a deletion of the Rubicon or adenoviral delivery of Atg7[202,251]. Recent evidence also indicates that impaired mitophagy may contribute to liver injury during progression of NAFLD and formation of megamitochondria[229].
Transition of NAFLD to NASH also implicates Kupffer cells. These cells, constitute 80%-90% of tissue macrophages in the body and are critical cells in liver inflammation[20]. They are the main site of NLRP3 inflammasome activation and production of the pro-inflammatory cytokines compared to hepatocytes and stellate cells[267,268]. Activation of the NLRP3 inflammasome plays an important role in the transition from NAFLD to NASH[269].
An earlier report demonstrated that cathepsin B, a lysosomalcysteine protease, is released in the cytosol in response to FFAs and that this redistribution of cathepsin B is present in the liver of patients with NAFLD related to disease severity. Importantly in a dietary mouse model of NAFLD, inhibition of Cath B significantly decreased steatosis, liver inflammation and insulin resistance[270].
These findings were recently elaborated in more detail as it was reported that cathepsin B and activation of the NLRP3 inflammasome are interconnected in a murine model of NASH but also in isolated Kupffer cells stimulated with palmitate. Expression of cathepsin B and activation of NLRP3 inflammasome were increase in NASH animals. Moreover, an inhibition of Cathepsin B decreased liver inflammation, ballooning, and the pro-inflammatory cytokines IL-1β and IL-18. In vitro stimulation of Kupffer cells showed identical results in inflammasome activation, expression of Cath.B and cytokine production before and after Cath.B inhibition. These results indicate that NASH pathogenesis probably depends in part to inflammasome activation which in turn is regulated by the activity of aprotease tightly connected to autophagy[271].
Additional supporting evidence for the role of autophagy in NASH pathogenesis is the fact that impaired autophagy in obese mice is critical for macrophage polarization. M2 macrophage polarization relies on energy provided by FFA oxidation, suggesting a potential implication of autophagy in this process. Macrophages change to a pro-inflammatory phenotype due to both increased M1 and decreased M2 polarization[132] with a resultant upregulation of liver inflammation, a prominent feature of NASH.
The situation is controversial when adipose tissue macrophages from obese mice are concerned. Increased rather than decreased autophagy of macrophages has been demonstrated in adipose tissue[272,273]. Another cathepsin mostly found in Kupffer cells seems to be implicated in NASH. Lysosomal cholesterol accumulation inside murine Kupffer cells leads to increased liver Cathepsin D activity which is related to liver inflammation[274]. Kupffer cell cathepsin D may therefore be an additional key player in hepatic inflammation of NASH[275]. The impairment of macrophage autophagy with aging may explain in part the increased prevalence of the metabolic syndrome and steatohepatitis of older age in humans[276,277].
The oxidative stress is also involved in the progression to NASH. Hepatocytes exposed to palmitate concentrations similar to those found in patients with the metabolic syndrome and NAFLD showed mitochondrial membrane permeabilization and production of ROS. Similarly, an inhibition of Cathepsin B ameliorated mitochondrial dysfunction and oxidative stress, indicating an additional mechanism of NASH progression[229,278].
Under normal conditions, damaged mitochondria are removed through mitophagy. In certain cases of NAFLD however mitophagy is defective and the oxidation of biomolecules by mitochondrial ROS starts a vicious cycle of increasing mitochondrial dysfunction and aggravation of hepatocellular oxidative damage. This ultimately leads to hepatic inflammation and liver failure[279,280], since impaired mitophagy triggers liver NLRP3 inflammasome activation in vivo and in vitro in isolated murine hepatocytes[38].
Impairment of autophagy in other liver sinusoidal cells may also participate in the progression of NAFLD to NASH. Decreased autophagy has been observed in the liver endothelial cells of patients with NASH or in mice with endothelial deletion of Atg5 and features of inflammation[180,190,281]. A very recent study has convincingly shown that impaired autophagy of liver endothelial cells (LSECs) occurs in NASH patients but not in simple steatosis. Deficiency in autophagy in LSECs induces endothelial inflammation ultimately leading to liver inflammation and fibrosis. This defective autophagy, in part due to inflammatory mediators of the portal blood, might well be one of the missing links of the progression of simple steatosis to NASH and cirrhosis[282].
A further mechanisms leading to NASH involves multivesicular bodies (MVBs), a form of endosomes, whose contents are transported into lysosomes[283]. The MVB-lysosomal pathway was shown to participate in the development of steatohepatitis through lysosomal degradation of Toll-like receptor 4 reported to be critical for the progression of NASH[284].
Finally a role of the chemokine CXCL10 in the development of steatohepatitis has been proposed. Upregulation of CXCL10 impairs autophagic flux decreasing thus autolysosome formation. Autophagic protein degradation is inhibited followed by the accumulation of ubiquitinated proteins with ultimate development of steatohepatitis[285].
The liver is the organ mostly responsible for ethanol metabolism. Oxidation of ethanol happens through three pathways namely alcohol dehydrogenase in the cytosol, cytochrome P450 (CYP2E1) in the ER and microsomes and the enzyme catalase in peroxisomes[286]. Ethanol oxidation also produces ROS, including superoxide anion, and hydroxyl radicals that may damage hepatocytes[287].
Ethanol induces autophagosome formation in the liver. Reduction of autophagy results in the accumulation of lipid droplets and apoptosis of hepatocytes[288]. On the other hand activation of autophagy by rapamycin attenuates steatosis and injury induced by a combination of ethanol and lipopolysaccharide[289].
Induction of autophagy by acute ethanol exposure is mediated through many mechanisms Ethanol-induced autophagy requires ethanol oxidation to acetaldehyde and ROS generation[290,291]. ROS activates autophagy by suppressing mTOR and proteasome activity[292,293] and inactivation of Atg4[294].
Oxidants differentially influence the activities of the proteasome (the other major pathway of protein degradation.) Proteasomes are reduced when autophagosomes are increased[295]. Proteasome inhibition further triggers ER stress activates autophagy through JNK activation. Ethanol may also suppress Akt and mTOR through the upregulation of PTEN[296,297]. Metals, like zinc, are also implicated in autophagy alterations after ethanol treatment[298].
A caution should be exercised on CYP2E1 ethanol oxidation as oxidative products resulting from the expression of CYP2E1 may in fact impair autophagy leading to lipid accumulation in the liver. In cells expressing CYP2E1, hepatocyte lipids and generation of ROS were increased by an inhibitor of autophagy and decreased when a stimulator of autophagy was used[299]. Similar results were found after acute alcohol in CYP2E1 knockout mice[291]. These findings also support the idea that autophagy protects against ethanol/CYP2E1-dependent hepatic injury.
It has also been shown that hepatic autophagy depends on the level of acetaldehyde produced during ethanol metabolism. Mice expressing the ALDH2 isoenzyme, clear acetaldehyde more rapidly and have increased autophagy and lower levels of hepatic triglycerides[300]. Cannabinoid receptor 2 can also induce macrophage autophagy to protect from alcoholic liver damage[301].
It should be stresses however that acute and chronic ethanol exposure may have different effects in liver autophagy[302]. Increased autophagosome formation and autophagy flux were shown in cultured hepatocytes after short term incubation with ethanol or in livers of mice after acute alcohol administration[288,302]. Enhanced autophagy parallel a higher hepatocyte nuclear content of TFEB, the main transcriptional regulator of genes involved in lysosome biogenesis[49,50].
Alcohol also has an effect on the transcription factor forkhead box O3a (FoxO3a) that modulates liver autophagy[303]. The activity of FoxO3a is largely controlled by multiple post-transcriptional modifications, including phosphorylation and acetylation[304]. Acute ethanol exposure increases nuclear translocation of FoxO3a inducing its dephosphorylation and acetylation.
However, results are not uniform for the chronic ethanol effect. Chronic ethanol administration (Lieber-DeCarli model) for 4 wk or 10 wk increased autophagosome numbers in murine livers, suggesting the induction of autophagy[305]. In another similar murine model, mice were given gradually increasing ethanol ethanol concentrations for 10 d and autophagic flux was reduced[302].
The discrepancy seems to be solved by the report that autophagy response was dependent on the alcohol concentration used. In a murine model on Lieber-DeCarli diet with different levels of alcohol for 4 wk, autophagy is increased by a lower dose of alcohol (29% of the caloric need), but decreased by a higher dose (36% of the caloric need). Liver injury was aggravated by further reduction of autophagy and attenuated by autophagy activation[306].
Earlier studies have also demonstrated that chronic alcohol exposure disrupts lysosome function[307]. Overall results have demonstrated that autophagy is suppressed in chronic alcohol consumption due to either the defect of lysosomal function and biogenesis from TFEB suppression[302,308] or to a reduction in AMPK activity and inhibition of autophagosome formation[309,310].
After ethanol-induced reduction of autophagy, there is accumulation of aggregated proteins and SQSTM1/p62, leading to activation of nuclear factor erythroid 2-related factor 2 (Nrf2) and damage to the mitochondria and cell death[309,311].
How the other autophagy-related transcriptional factors, such as TFEB and farnesoid X receptor (FXR) are interconnected with FoxO3a in the expression of autophagy genes is unknown. Moreover, how ROS generation in acute or chronic alcoholic condition systematically affects the mTORC1 activation or TFEB translocation is unclear.
Autophagy is also protective against CYP2E1-dependent liver lesions in a chronically ethanol-fed murine model[312]. Autophagy in ALD can be further affected by additional factors identified in various experimental models. Augmenter of liver regeneration (ALR) is a factor that can promote liver growth. It was reported to protect mice from ethanol-induced liver injury through inhibition of mTOR and therefore activation of autophagy[313]. Moreover an interesting recent study used many genetic models of autophagy impairment, with different functional levels and different alcohol regimens. Deficiencies of either Atg7 or Atg5 demonstrated variable responses to ethanol feeding according to the timing of autophagy dysfunction, the gene being affected, and the alcohol scheme used[314].
It should be stressed that in acute alcohol administration, ethanol-induced autophagy may protect the liver by three basic mechanisms namely mitophagy[80,102,315,316], lipophagy[72,293,317] and clearance of Mallory-Denk bodies by proteophagy[265,318,319].
However, chronic alcohol exposure impairs autophagy and lipophagy[308,320] most likely due to the activation of mTOR signaling and a decrease in lysosomal biogenesis. Administration of the mTOR inhibitor Torin- 1 restores lysosomal biogenesis and attenuates liver lesions[308]. An additional pathway through which chronic alcohol exposure could reduce liver autophagy is the inactivation of the guanosine triphosphateRab7 and reduction of dynamin 2 activity leading to depletion of lysosomes and inhibition of hepatocyte lipophagy[320,321].
Ethanol Induced steatosis activates mitophagy by elevating PINK1 expression on mitochondria[305]. PINK1-dependent mitophagy was correlated with the mitochondrial expression of Parkin and the level of an indicator of oxidative mtDNA damage[322-325]. Mitophagy has a dominant role in protection of the hepatocyte from alcohol-induced hepatic injury as evidenced by a report that enhancement of mitophagy by quercetin, a natural flavonoid, attenuated ethanol-induced mitochondrial damage[326].
Regulation of mitophagy is related to three receptors namelyFUN14 domain containing 1 (FUNDC1), BCL2 interacting protein 3 (Bnip3), and Parkin[327].
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is a newly described housekeeper of liver mitochondrial fission. DNA-PKcs is overexpressed in murine livers after exposure to ethanol and was positively correlated with steatosis, mitochondrial damage andfibrosis. On the other hand this over expression repressed FUNDC1-required mitophagy[328].
An additional significant point is the effect that ethanol might have on the different sinusoidal cell subpopulations. There is strong evidence that autophagy in macrophages is crucial to protect the liver from ethanol-induced damage. Investigations were mostly performed in macrophage specific deletions of either Atg7 or Atg5. The cannabinoid CB2 receptors of macrophages were found to have a protective rolein ALD, which was abrogated by Atg5-deletion in macrophages[301]. Increased mortality in Atg5 deleted mice was also demonstrated after chronic ethanol feeding plus LPS challenge[329]. Similar findings were reported after Atg7 deletion[330]. Both studies demonstrated an activation of the inflammasome and an augmented IL-1 production.
In contrast to hepatocytes and macrophages the effect of autophagy in hepatic stellate cells after ethanol exposure has not been clarified. A recent study in immortalized rat stellate cells demonstrated that autophagy could contribute to ethanol-induced stellate cell activation[331]. Induction of fibrosis by alcohol in current murine models is not feasible unless accompanied by steatosis induced by a high-fat diet[332].
Most autophagy studies in ALD are focused on the involvement of macroautophagy. Recent evidence however indicates that CMA is also important in alcoholic liver disease through the CMA negative regulator sorting nexin 10 (snx10). Snx10 knockout mice fed with Lieber-DeCarli diet were resistant to alcohol-induced liver injury associated with an increase of lysosome-associated membrane protein 2A (LAMP2A) and CMA activation through inhibition of the enzyme Cathepsin A which is responsible for LAMP2A degradation[333]. Deficiency therefore of a CMA negative regulator, protects animals from ALD. Deficiency of another CMA negative regulator, Lipocaline-2 (LCN2), also maintains hepatic CMA activity in murine livers after chronic alcohol administration[334] verifying the idea that impaired CMA may be responsible at least in part in alcohol-induced liver injury.
Involvement of miRNAs is an additional factor in the regulation of autophagy in ALD that has emerged from recent evidence. Several miRNAs were reported to alter autophagy and alcoholic steatosis[335]. miR-26a ameliorates alcohol-induced acute liver injury by two MAPKs inhibitors thus inducing Beclin-1 expression and autophagy[335]. Another report provided evidence that miR-155 is a mediator of alcohol-related exosome production and autophagy impairment in both hepatocytes and macrophages[336]. Deletion of miR-155 protected mice from alcoholic steatosis and inflammation. Interestingly in this study serum levels of exosomes were increased in ALD patients and alcohol exposed mice, whereas miR-155 deficient mice had significantly reduced exosome release from both hepatocytes and Kupffer cells. It was suggested therefore that autophagy is an atypical promoter of exosome release in ALD.
Clinically important observations indicate that withdrawal of ethanol from ethanol-fed rats resolves steatosis[337] suggesting that removal of ethanol oxidation and restoration of lipophagy may be the mechanism of steatosis resolution observed in humans after ethanol abstinence[338,339]. Informative reviews of autophagy in ALD were recently published[90,181,182,340-342].
In view of the fundamental role of lipophagy in the pathogenesis of ALD, it is not surprising that pharmacological inducers of lipophagy like carvamazepine, rapamycin, resveratrol and simvastatin were tested in alcohol-fed animals with a resultant attenuation of liver lesions. By contrast chloroquine exacerbated hepatic steatosis[312,343,344]. Recently plant-derived agents were also used to activate lipophagy. Thus, corosolic acid[345], quercetin[346] and Salvianolic acid A[347] all had a favorable result on alcohol-induced liver lesions activating lipophagy through different pathways.
Summarizing, it is evident that whether ethanol causes an increase or decrease of autophagy depends on the duration of ethanol consumption/exposure, the amount of alcohol given, and the manner in which it is administered[290,302]. Moreover, lipophagy and mitophagy cannot act as defensive mechanisms in the long term as they do in acute ethanol consumption as they are inhibited by chronic alcohol exposure[102,348].
In the past decade, hepatic autophagy has been implicated in viral infection with either hepatitis B (HBV) or hepatitis C (HCV).
Recent studies have shown that autophagy is involved in the life cycle of Hepatitis B. Inhibition of autophagosome formation could reduce HBV production, while stimulation of autophagy could significantly contribute to HBV production[349,350].
However, the mechanism by which HBV activates autophagy is not clear. Previous reports have implicated either the HBx[351,352] the large HBsAg protein[353] or a mutant with a deletion in the preS2 region[354,355] as inducers of ER stress which in turn increases autophagy.
In contrast it was shown that HBx does not play a significant role in the induction of autophagy compared to the small HBsAg protein also increasing autophagy via the induction of ER stress. An HBV genome unable to express small HBsAg does not activate autophagy[356]. To reconcile the discrepancy, it has been suggested that autophagy can be stimulated both by HBx and the small surface HBsAg protein through upregulation of beclin-1 expression[357,358]. In addition HBx induces autophagy through its effect on the cytoplasmic high-mobility group box 1 (HMGB1), identified as a a positive regulator of autophagy. HBx binds to HMGB1 and triggers autophagy in hepatocytes[359]. This observation may be clinically relevant. Spontaneous and induced autophagy of peripheral Treg cells from 98 patients with chronic hepatitis B were assessed[360]. No difference of spontaneous autophagy was found between patients and normal controls but induced autophagy was significantly higher in patients. It was also related to HMGB1 as it was significantly decreased when HMGB1 was blocked with a neutralizing antibody.
HBx further impairs lysosomal acidification with a final result the accumulation of immature lysosomes. Moreover immature lysosomal hydrolase cathepsin D was shown in human liver tissues with chronic HBV infection suggesting that a repressive effect of HBx on lysosomes may be responsible for the inhibition of autophagic degradation[350]. Interestingly, although HBV could impair lysosomal acidification it was unable to induce autophagic protein degradation, due to the inability of HBV to increase the sequestration of proteins destined for degradation by autophagy[350]. Therefore, it is usually stated that HBV induces incomplete autophagy. In addition, it was clearly shown that HBV specifically targets damaged mitochondria and mitophagy. Either the whole HBV genome or HBx alone were able to induce Parkin-mediated mitophagy[361,362]. In addition, HBx-induced autophagy inhibited mitochondrial apoptosis increasing the survival of HBV DNA-transfected cells[349]. Another clinically important observation is that different HBV genotypes have a variant effect on autophagy. HBV genotype C was a more potent inducer of autophagy than HBV genotype B. HBV-C is associated with more severe disease than HBV-B but however attractive such an association between autophagy and severity of liver disease may be, it has to be verified[363,364].
It is important to realize that many viruses, including HBV, have developed strategies to hijack autophagy to benefit their replication and dissemination[356,365,366]. So far, HBV is the only DNA virus known to exploit autophagy for its own replication as it is RNA, but not DNA viruses, that commonly use autophagic function to promote replication[367].
HBV infection induced the early-stage formation of autophagic vacuoles increasing the PI(3)K enzyme activity to promote HBV DNA replication. HBx can directly bind and activate the PI3KC3 complex[368,369]. Ablation of Atg5 has been shown to inhibit autophagy and impair nuclear localization of the HBV core protein. HBV DNA level in sera was decreased by more than 90% accompanied by practically undetectable levels of the HBV DNA replicative intermediate in the liver[370].
Autophagy was responsible for the degradation of an oncogenic microRNA-224 in the liver of HBV patients with hepatocellular carcinoma (HCC) and HBx-transgenic mice. In HCC patients, the combination of low-Atg5 expression and high miR-224, was significantly correlated with a poor overall survival rate[371]. The list of the mechanisms used by HBV to subvert autophagy and the detrimental consequences in the liver is by no means complete as new factors are constantly reported including release of pro-inflammatory cytokines and chemokines and inhibition of neutrophil extracellular trap[372-375].
Further evidence of autophagy subversion by HBV was recently reported. In HBV-replicating hepatocyte cultures, the silencing of Atg5, Atg12, and Atg16L1, interfered with viral core/nucleocapsid (NC) formation/stability and significantly reduced virus yields. It was further demonstrated that a covalent conjugation of Atg12 to Atg5 was essential for HBV replication. In addition the virus required Atg10 and Atg3 which are necessary for Atg5-12 conjugation. Deletion of Atg10 and Atg3 decreased HBV yields, while Atg3 overexpression increased virus production. HBV was associated with the Atg5-12/16L1 via interaction of HBV core protein with the Atg12 unit of the complex. Subsequent autophagosome maturation events were not necessary for HBV replication. These data indicate that HBV subverts early, non degradative autophagy components avoiding thus autophagosomal destruction[178,376,377].
Death receptors of TNFSF10 (tumor necrosis factor superfamily member 10) participate in the immune defense against several viruses by promoting apoptosis. HBx impairs TNFSF10 receptor signaling through autophagy mediated lysosomal and not proteasomal degradation. Importantly a significant reduction of the protein TNFRSF10B was demonstrated not only in cell lines but also in the liver of chronic HBV patients[378].
It was very recently reported that the hepatitis D virus also utilizes autophagy to assist its life cycle as it increases autophagosome accumulation and impairs autophagic flux. Both the small HDAg and large HDAg proteins are capable to disturb the autophagy machinery, in particular the proteins Atg7, Atg5, and LC3 involved in the early elongation stage of autophagy. Unexpectedly, deletion of Atg5 and Atg7 reduced the intracellular HDV RNA level in hepatocyte cell lines without an effect on HDV secretion[379]. Reviews of autophagy in HBV have recently been published[366,380].
Reported data have shown that HCV could induce autophagy to support its own replication[381,382]. Several mechanisms for HCV induction of autophagy have been investigated using hepatocyte cell lines[383,384]. HCV infection initiates the formation of phagophores after induction of the localization of Atg5 to the ER. Phagophores fuse to form autophagosomes. HCV-induced autophagosomes were further reported to be required for viral RNA replication as the autophagosomal membrane provided a platform containing HCV NS5A, NS5B, and viral RNA for replication[385-387] but subsequently HCV blocks the fusion of autophagosomes and lysosomes through Rubicon overexpression. As a result autophagosomes accumulate and HCV RNA replication and assembly of infectious virions[385,388,389,390,391] are supported.
However, several studies have contradicted the need for co-localization of viral proteins in the autophagosomal membrane suggesting that this is not a necessity for viral replication[392-395].
Autophagy favors HCV replication with an additional mechanism. The entire autophagic process may be manipulated leading to the suppression of the HCV associated innate antiviral response[393,396]. After silencing different Atgs, HCV viral infectivity was suppressed in parallel with an upregulation of interferon-stimulated gene expression[390]. Moreover, HCV seems to activate autophagy to degrade the tumor necrosis factor receptor -associated factor 6 (TRAF6), thus subverting innate host immunity[389,397-399]. HCV induced unfolded protein response strongly activates autophagy to sustain viral replication through inhibition of cellular apoptosis[396]. Different HCV genotypes may have variable influence on autophagy[391,400].
HCV was also found to selectively activate lipophagy to counteract the HCV induced lipid abnormalities. This may be clinically important as the levels of autophagy in the liver of chronic HCV patients were inversely correlated to steatosis[401]. Inhibition of autophagic degradation of lipophagy may account for the characteristic occurrence of hepatic steatosis in chronic HCV infection. Mitophagy is also selectively activated via the PINK1–Parkin axis in infected cells, thereby promoting HCV viral RNA replication[361,402]. Virus-activated mitophagy further attenuates apoptosis and favors persistent viral infection[403]. In agreement with this finding, the viral non-structural protein 5A (NS5A) was shown to disrupt mitochondrial dynamics, thus increasing ROS production and mitophagy[404].
On the other hand, the viral core protein interacts with Parkin inhibiting its translocation to mitochondria. Mitophagy is suppressed and mitochondrial injury of infected hepatocytes is sustained and viral persistence is maintained[405].
Syntaxin 17 is an autophagosomal protein required for the fusion of autophagosomes with lysosomes and also the release of HCV. The amount of syntaxin 17 was reduced in HCV-replicating cells indicating that HCV impairs the late stages of autophagy affecting the equilibrium between the release and the lysososomal degradation of viral particles[406].
Recently CMA was also demonstrated to be activated by HCV leading to degradation of IFN-alpha receptor-1[407]. Moreover the HCV NS5A was found to interact with Hsc70, recruiting Hsc70 to hepatocyte nuclear factor 1 alpha thus targeting HNF-1α for CMA degradation[408]. Taken together these studies indicate that HCV induced CMA also facilitate HCV replication.
However, an opposite less permissive effect of the manipulation of autophagy by HCV has been suggested as a result of recent studies. Atg10 is critical for autophagy as it promotes the Atg5-Atg12 complex formation. Two isoforms of the Atg10 protein were described, namely Atg10 (a longer one) and Atg10S. They have a similar amino acid sequence except for an absence of a 36-amino acid fragment in Atg10S. Yet they differ in their effects on HCV genome replication. Atg10 with deleted or mutated two cysteins, (Cys44 and Cys135) could trigger the expression of anti-HCV immunological genes combating the HCV replication[409,410].
Taken together these results indicate that autophagy is required for initiation of the HCV replicative phase but not for further replication[393]. However this might not be entirely true, as chloroquine an inhibitor of lysosomal acidification inhibits HCV replication offering an additional evidence for the permissive role of autophagy in HCV infectivity in the late phase[411].
Autophagy may additionaly be involved in HCV replication through the regulation of the exosomal pathway[390] and apolipoprotein transport[412], both critical steps in the egress of the HCV virion. The virion is associated to apolipoprotein E (ApoE) and its infectivity is enhanced. Autophagy has a central role in the trafficking of ApoE in HCV-infected cells leading to partial autophagic degradation of ApoE, but also to the interaction between ApoE and the viral protein E2 to increase the production of infectious viral particles[412].Molecular details of how HCV is using autophagy to its own advantage were recently published[380,413].
In summary, the life cycles of HBV and HCV in liver cells can be subdivided into 7 steps: Endocytosis, uncoating, genome replication, translation, envelopment, assembly and release. Both HBV and HCV drive autophagy largely by the ER stress response resulting from uncontrolled translation of viral proteins[414-416]. In addition HBx modulates autophagy for the benefit of HBV replication[357], while multiple HCV proteins including p7, NS3/4A and NS4B, modulate autophagy by direct or indirect association with moieties of the early autophagy machinery in favor of its replication[417-419]. Pharmacological or genetic manipulation of autophagy may limit the viral yield[183,369,420], making autophagy a feasible target for HBV and HCV treatment.
The liver responds to practically any insult with only a limited number of pathological lesions: Hepatitis (hepatocyte death), cholestasis, fibrosis-cirrhosis or a combination of the three. Autophagy participates in all liver pathological responses.
Liver fibrosis is a complex and dynamic cellular process implicated in the evolution of the majority of chronic liver disease towards cirrhosis. Most review articles have broadly concentrated on the role of autophagy in liver diseases, with restricted information on cell types implicated in liver fibrosis. Not unexpectedly, most research has focused on hepatic stellate cells (HSCs) and myofibroblasts, because they are the central elements in extracellular matrix production[421]. However, other liver cells, including hepatocytes, macrophages, sinusoidal endothelial cells (LSECs), infiltrating immune cells and the so-called ductular reaction (DR) are also important[422,423]. DR significantly correlates with the degree of fibrosis and involves cholangiocyte-like cells that dominate an interplay of extracellular matrix and inflammatory infiltrate[424-427].
The fundamental event in fibrosis is the transformation of hepatic stellate cells into myofibroblasts and this is closely related to autophagy. Typical autophagosomes that contained LDs were found in cultured HSCs indicating a connection of liver fibrosis and lipid autophagy[428]. Increasing evidence supports the notion that inhibition of lipophagy in hepatocytes reduces HSC activation and fibrosis progression[429,430]. Inhibition of the activation of HSCs and the formation of autophagosomes have been reported and these seem to be connected with the downregulation of transforming growth factor beta 1/Smads pathway as an increase in TGFb/Smad3 Leads the transcription of Beclin-1, which is a critical player in the autophagy process[431-433].
In rat-derived HSCs, cytoplasmic LDs are degraded followed by fibrogenic genes expression. Moreover induced lipid accumulation by an alkaloid, was associated with quiescent HSCs due to autophagy blockade[434]. Inhibition of autophagy by chloroquine improved CCl4-induced liver fibrosis affecting the activation of hepatic stellate cells as expected[435]. On the other hand, dihydroceramide an inhibitor of autophagy promoted the progression of liver steatosis to fibrosis[436]. Similarly, inhibition of YAP degradation also led to liver fibrosis[113].
In addition, it has been suggested that the IL-17A/STAT3 signaling pathway is important in the evolution of liver fibrosis through suppression of hepatocellular autophagy since neutralization of IL-17A promotes the resolution of experimental fibrosis[437].
Based therefore on current evidence, it has been stated that autophagy at least in murine hepatocytes is a selective survival mechanism through clearance of excessive fat leading to attenuation of lipotoxicity[438]. This is certainly not the case for HSCs autophagy where lipid droplets are digested to supply energy for the activation of HSCs, promoting thus liver fibrosis. Non specific inhibition of stellate cell autophagy or specific inhibition of Atg5 or Atg7, blocked HSCs activation[439-441]. Lipophagy in HSCs is induced by ER stress[442] and is mediated through Rab25 in a ROS dependent manner as antioxidants were effective in stopping autophagy[87]. In agreement with experimental data, clinical research found that cirrhotic patients had significantly increased levels of several autophagy- related genes compared with non cirrhotics accompanied by increased maturation of lysosomal cathepsin D[85]. Furthermore, serum lipids were evaluated in patients with cirrhosis of viral etiology and compared to non cirrhotics. Low serum lipids were found in HCV and HBV cirrhosis which were negatively correlated with lipophagy[443].
Micro-RNAs interfere with the activation of stellate cells. miR-16 inhibits the expression of guanine nucleotide-binding -subunit 12 (G12) which is overexpressed during fibrogenesis and facilitates Atg12-5 formation, thus activating stellate cells[444]. Also miR-181-5p transferred to mouse HSCs via exosomes from engineered adipose derived stem cells led to inhibition of fibrosis[445].
Several signals can induce autophagy in HSCs[180], including hypoxia-inducible factor-1alpha[446], transforming growth factor 1[447], as well as the danger-associated pattern molecule high-mobility group box-1 (HMGB-1)[448]. Additional signals like ROS-JNK1/2 and the XBP1 arm of the Unfolded Protein Response have also been identified as necessary requirements of HSCs activation through autophagy[449,450]. TGF-β1 has also been reported to mediate autophagy[440]. Similarly, HSCs in cell culture with depleted Atg2A fail to spontaneously trans-differentiate[451]. Quercetin attenuated hepatic fibrosis in mice through inhibition of hepatic HSC activation and autophagy[452].
Selective activation of mitophagy in HSCs also favors fibrosis. PM2.5 is an air pollutant that activates HSCs and initiates liver fibrosis. This is due to increased ROS production and induction of mitophagy through activation of the Pink1/Parkin pathway[453]. In contrast, inhibition of mitophagy was shown to promote inflammation[454] due to dissemination of inflammatory signals from HSCs production of inflammatory cytokines[455]. However very recently it was reported that selective inhibition of mitophagy in macrophages attenuates fibrosis. Mice Kupffer cells from CCL4-induced acute injury showed increased ROS production, activated mitophagy and increased TGF-β1 secretion. T-cell immunoglobulin domain and mucin domain-4 (TIM-4) interference in Kupffer cells inhibited Akt1-mediated ROS production and decreased mitophagy and TGF-β1 secretion through suppression of PINK1/Parkin, to ameliorate CCl4-induced hepatic fibrosis[456]. Seemingly in disagreement with this notion, is the finding that the autophagic proteinp62/SQSTM1, a negative controller of HSC activation is downregulated in trans-differentiating HSCs associated with hepatocellular carcinoma. P62 ablation increases fibrogenesis but this is not related to autophagy but rather to the reduction of p62-dependent activation of the vitamin D receptor (VDR) and the resultant loss of repression of HSC by VDR agonists[457,458].
Even in HSCs the characterization of autophagy as a double-edged sword has been justified. A novel molecular mechanism of selective autophagy in HSCs indicates that autophagy may also protect from liver fibrosis. The RNA-binding protein ELAVL1/HuR plays a crucial role in regulating ferroptosis in liver fibrosis. ELAV1 enhances ferritinophagy leading to ferroptosis of HSCs and attenuation of liver fibrosis[459]. Despite this report, most existing evidence indicate that activation of HSCs autophagy is pro-fibrogenic, therefore a selective block of autophagy in fibrogenic cells might be an attractive future anti-fibrotic therapy[90].
The opposite seems to happen in hepatic macrophages[55] where activation of autophagy is anti-fibrogenic[460]. Mice macrophages with specific deletion of atg5, secreted increased levels of ROS-induced IL-1A and IL-1B. In addition, liver myofibroblasts incubated with the conditioned medium of Atg5(-/-) macrophages expressed increased pro-fibrogenic genes. Attenuation of fibrosis was achieved after IL-1 neutralization indicating that IL1A/B are critical mediators of the profibrotic effects of autophagy inhibition in macrophages[461-463]. Autophagy in Kupffer cells is counteracted by the enzyme monoacylglycerol lipase catalyzing the production of arachidonic acid leading to inflammatory macrophage activation and fibrosis[464].
On the other hand deletion of Atg7 in sinusoidal endothelial cells (LSECs) demonstrated that the selective loss of their autophagy led to cellular dysfunction and decreased intrahepatic nitric oxide. Impairment of autophagy after CCL4-induced acute liver injury in rats, also impaired handling of oxidative stress by LSECs and amplified liver fibrosis[465].
Similarly, autophagy defective sinusoidal endothelial cells (LSECs) as demonstrated in patients with NASH favor advancement of fibrosis[282]. At the same time, even excessive autophagy activation may lead to caveolin-1 degradation, thus worsening the LSECs defenestration and ultimately promoting fibrosis[466]. Therefore, any dysregulation of autophagy in LSECs may aggravate liver fibrosis[467].
An elegant immunofluorescence study of cirrhotic livers linked autophagy with an additional population of fibrogenic cells other than HSCs, the reactive ductular cells (RDC) which were characterized as cholangiocyte-like epithelial cells positive for cytokeratin 19[85]. They are responsible for ductular reaction (DR), a common response to various insults of the liver implicated in the pathogenesis of cirrhosis[432]. Administration of chloroquine, reduced the expression of CK19 positive RDC and blunted liver fibrosis[86]. DR parallels HSC activation in many liver diseases[430]. Reactive ductular cells secrete soluble pro-fibrogenic factors targeting HSCs and myofibroblasts[468]. Recently it was demonstrated that in cirrhotic human livers, RDCs with activated autophagy also had upregulated expression of TGF and fibroblast specific protein-1[469] making autophagy a necessary requirement during the DR process. The role of autophagy in liver fibrosis is therefore complex and the end result depends on the cell population involved. In general, HSCs and RDCs have a pro-fibrogenic effect. On the contrary, autophagy counteracts fibrogenesis acting in hepatocytes, macrophages and LSECs[470].
The role of autophagy in tumor cell biology has not been fully elucidated. Autophagy has both pro-and anti-tumorigenic roles. For example, it can either inhibit inflammation acting as an anti-oncogen or protect tumor cells from ROS damage acting as a pro-oncogen[471,472].
Opposing effects have been reported. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis[473]. On the other hand, Ras-induced expression of two proteins Noxa and Beclin-1 promotes autophagic cell death, limiting thus the oncogenic potential of deregulated Ras signals[474]. Drugs like ursodexocycholic acid can efficiently eliminate resistant to other drugs cancer cells through induction of autophagic death[475].
HCC is one of the most common types of liver cancer[476]. Most of the HCC cases are accompanied by cirrhosis that results from long-standing chronic inflammation due to viral hepatitis or non-viral etiologies including heavy alcohol intake, NAFLD, autoimmune hepatitis, primary biliary cholangitis, and hemochromatosis[477].
Mice with impaired autophagy are unable to develop HCC even after of strong challenge. This was related to the induction of tumor suppressors like p53[478]. However, after initiation of HCC, the presence of autophagy is required to degrade tumor suppressors promoting thus the development of HCC[86]. Both macroautophagy and CMA are implicated as a double edge sword in liver tumorigenesis[479].
Autophagy has a dual role in hepatocellular carcinoma (HCC). It is an anti-cancer mechanism in the dysplastic stage of HCC initiation, while it favors HCC development and confers resistance to treatment[480,481]. This is possibly due to the maintenance of mitochondrial integrity and protection of cells against oxidative stress during HCC initiation, followed by the downregulation of tumor suppressors to promote the development of HCC[86,482].
In a study of 156 HCC patients increased levels of the autophagy marker LC3B are associated with a dismal prognosis[483]. Higher levels of LC3-II were associated with lymph nodes metastasis, higher vascular invasion and reduced 5-year survival[484].
Macroautophagy may also have an anti-oncogenic function, as reduction of either Atg5 or Atg7 Levels lead to appearance of multiple liver tumors[485]. Similarly, low levels of autophagic proteins and activity are associated with bad prognosis of human HCC[486,487]. Beclin-1 Levels are lower in HCC tissue samples compared to normal tissue from the same patient. Beclin-1 expression was studied in 300 HCC patients. A correlation with disease-free survival and overall survival was found only in the Bcl-xL+ve patients. It was suggested therefore that a synergy of defective autophagy and altered apoptotic activity lead to tumor progression and reduced survival[488].
Inhibition of autophagy leads to the accumulation of SQTSM1/p62. Accumulation of p62 on the one hand may protect from HCC initiation as it blocks the antioxidant functions of nuclear factor erythroid-2- related factor 2 (Nrf2)[489-492]. On the other hand, accumulation of p62 also contributes to carcinogenesis through persistent activation of Nrf2[493]. Nrf2 expression promotes the development of HCC[493]. Deletion of p62 in autophagy defective livers counteracts tumorigenesis. Therefore an accumulation of p62 is partly responsible for the increase in hepatic tumors, via the activation of Nrf2[492-494]. The activation of Nrf2 turns glucose and glutamine into anabolic pathways supporting tumor cell proliferation[176,495]. In addition, autophagy inhibits malignant transformation in the liver through Yes associated protein 1 (YAP1) degradation, a protein with a crucial role in hepatic oncogenesis[113,496].
Aberrant activation of the Wnt/β-catenin signaling is another critical pathway in the onset and development of HCC. A recent study reported that the Wnt/β-catenin inhibitors exert anti-tumor effects on HCC cells by regulating autophagy[497]. However this is in disagreement with a previous report where interfering with Wnt secretion in HCC cell lines does not affect autophagy or the level of β-catenin signaling despite cell growth suppression indicating that other mechanisms might underlie the growth-suppressive effect[498].
Furthermore, the activation of autophagy was shown to mediate inhibition of proliferation and induction of apoptosis of hepatoma cell through several mechanisms[499-506]. The induction of autophagy by concanavalin A or different chemotherapeutic drugs in murine livers inhibit hepatoma cell growth and prolongs survival[507-519]. On the other hand suppression of autophagy was reported to enhance the susceptibility of hepatocellular cancer cells towards a variety of chemotherapeutic agents[108,520-529].
Several microRNAs (mirRNAs) have been implicated in HCC tumorigenesis. miR-204 reduces tumor autophagy in HCC[530]. Moreover autophagy degradation of miRNA-224 suppressed the growth of HBV-related HCC[371], while miR-375 which is downregulated in HCC was reported to inhibit autophagy by decreasing the expression of Atg7 and autophagic flux. Up-regulation of miR-375 inhibits mitophagy of HCC cells, reduces the elimination of damaged mitochondria, and decreases cell viability[99]. miR-26 could inhibit autophagy and enhance chemosensitivity of HCC cells[531].
LncRNAs are another set of ncRNAs with a length exceeding 200 nucleotides without translation into proteins[109]. Several lncRNAs, like Hnf1a-as1, Hotair and Hulc promote autophagy and function as oncogenes in HCC[106-109].
The role of mitophagy and lipophagy is also important in HCC growth acting as a double edge sword. Increased mitophagy by concanavalin A, adriamycin or curcumin was shown to suppress hepatoma cell growth[507,510,517,530] while melatonin increased the sensitivity of human hepatoma cells to sorafenib by triggering mitophagy[532]. A recent study also demonstrated that inhibition of inflammasome activation and induction of mitophagy suppressed HCC growth[533]. On the other hand it has been demonstrated that increased mitophagy may facilitate HCC cell survival either through ROS production or attenuation of p53 activity[534,535].
Lipophagy can also act both ways. On the one hand, it can allow tumor cells to have access to a supply of energy critical to their growth[536] and on the other hand, lysosomal acid lipase, the lipase that facilitates lipophagy, exhibits tumor suppressor activity[537]. Lipophagy was also reported to induce apoptosis in vitro, via induction of ER and mitochondrial stress[538]. CCAAT enhancer binding protein a, a protein that is upregulated in HCC patients, increases resistance to energy starvation and favors carcinogenesis through lipophagy[539].
In addition to the general characteristics of autophagy implication in HCC, there are certain points to be mentioned in specific liver disease associated HCC. As mentioned before, autophagy is activated by the HBx protein[357,369] and this may be related to HBV carcinogenesis. Increased autophagosome formation by HBx was accompanied by decreased degradation of LC3 and SQSTM1/p62 and greatly impaired lysosomal acidification and accumulation of immature Cathepsin D. These data may indicate that repression of lysosomal function by HBx could be important for the initiation and progress of HBV-associated HCC[350].
CMA and cancer metabolism are also interconnected. Once malignant transformation occurs, CMA activity is significantly increased in cancer cells so that the new metabolic requirements are maintained[64]. Blockade of CMA which is upregulated in several cancers reduces progression and metastatic potential of solid tumors because the characteristic increased rates of aerobic glycolysis are reduced in a p53-dependent manner[540]. Macroautophagy and CMA seems to be interconnected and often substitute for one another as in the case of HCC. Under physiological conditions there is no expression of p62 in normal livers pointing to macroautophagy as the main mechanism facilitating cell survival. However in a recent study of 46 cirrhotic livers it was shown that p62 was increased indicating an impairment of macroautophagy, but LAMP-2A and heat shock protein 70 were uniformly increased indicating that an upregulated CMA was trying to compensate for the reduced macroautophagy and therefore promote HCC survival. Moreover, hydroxychloroquine inhibition of lysosomal degradation led to induction of the tumor suppressor p53 and promotion of apoptosis[541]. HCV is also an inducer of HCC. During HCV infection, increased cellular stress has been reported. Severe stress promotes Nrf2 transcription which in turn is responsible for CMA activation resulting in the suppression of hepatic innate immunity and possible degradation of tumor suppressors. The subsequent oncogenic cell programming initiated by a cytoplasmic virus like HCV, has been recently described in detail[542].
Defective autophagy is linked to MAFLD-related HCC, because the accumulated p62/SQSTM1, induces the oncogenic NF-κB activity while retained damaged mitochondria and produced ROS to damage cellular DNA[543]. A novel mechanism was recently reported in ethanol induced liver disease and HCC. Tumor necrosis factor-α-induced protein 8 (TNFAIP8) has been associated with tumor progression in several cancer types including the initiation of HCC. TNFAIP8 induced autophagy in liver cancer cells through blocking of AKT/mTOR signaling and direct interaction with ATtg3-Atg7 proteins. This mechanism is operative in alcohol related liver disease in mice and humans but not in high-fat-fed obese mice or patients with MAFLD[544]. Details of the molecular mechanisms of autophagy in both protection and promotion of HCC were recently published[545-547].
An additional aspect of HCC biology where autophagy plays an important role is the involvement of tumor-associated macrophages and tumor microenvironment. They are polarized after implication of sensing factors from tumor environment and autophagy[130,548]. Deficiency of TLR2 decreased the liver production of TNFα, IFN gamma and IL1a/b accompanied by reduction of autophagy flux and increase in oxidative stress and p62 aggregates in liver tissue. These changes were associated with increased carcinogenesis and progression of HCC[549]. Enhancement of autophagy in tumor-associated macrophages leads to M1 polarization which reduces tumor progression while M2 polarization is permissive for tumorigenesis[550]. The mTOR-TSC2 pathway, a key negative regulator of autophagy, is crucial for macrophage polarization since its activation leads to M2 phenotype. It was recently shown that the coagulants tissue factor (TF) and factor VII (FVII) produced in tumor microenvironment, are implicated in HCC growth promotion by suppression of autophagy mediated through mTOR activation and Atg7[551].
In view of the variable functions of autophagy, there should be an individualized approach of autophagy manipulations for HCC treatment. Thus, various lysosomal inhibitors including chloroquine and hydroxychloroquine have been used as treatment either as sole agents or in combinations with other treatment modalities in a variety of murine HCC models[523,552,553]. Interference with autophagy may be a sound therapeutic option for the treatment of HCC[554,555]. Based on the fact that autophagy is upregulated in metastatic HCC[556] use of autophagy inhibitors like chloroquine and hydrochloroquine in combination with other drugs may be a better option for treating metastatic HCC in humans. A combination of a number of drugs with autophagy inducers have been used to target cancer cells. A combination of percutaneous transarterial chemoembolization with chloroquine, was associated with increased tumor cell necrosis and apoptosis[557] and might counteract the presence of residual hepatocellular carcinoma cells[558,559]. Sorafenib, a multikinase inhibitor approved for HCC treatment, induces autophagy[560] and data show that a combination with autophagy inhibitors increase tumor response[537,561].
Xenografts in nude mice are widely used models of cholangiocarcinoma (CCA). Activated autophagy has been reported in tumor cells from such a model and in specimens from CCA patients[562]. LC3B, Beclin 1, and p62/SQSTM1 expressions were additionally found to be increased at the initial stage of the multistep cholangiocarcinogenesis[563]. However, a lower Beclin 1 expression was associated with metastatic lymph node disease and poor survival of patients with intrahepatic CCA[564,565]. Apoptosis was induced in cholangiocellular cell lines and tumor development was suppressed in a mice xenograft model after interference with autophagy[562]. Similarly, suppression of autophagy by chloroquine increased the chemosensitivity of cisplatin-treated CCA cells[566] and increased apoptosis of CCA cells through ER stress[567]. Chloroquine blockade of autophagy inhibited the tumor growth in Kras/p53 intrahepatic CCA[568,569]. CCA is extremely resistant to chemotherapy. 5-fluorouracil (5-FU) induced autophagy in CCA cells[570] while autophagy inhibition by capsaicin was followed by repression of malignant cell growth[570], indicating that autophagy may be implicated in the multidrug resistance of this tumor. Autophagy was also induced after incubation of CCA cells with the sphingosine kinase 2 inhibitor, ABC294640. Inhibition of autophagy by chloroquine potentiated ABC294640-induced apoptosis[571]. Modulation of autophagy therefore may be helpful in CCA treatment.
Autophagy is also implicated in other types of liver injury like the inherited metabolic diseases. Alpha 1-antitrypsin deficiency is the most extensively studied. Alpha 1-AT is a glycoprotein inhibitor of destructive neutrophil proteases[572,573]. Several naturally occurring mutants of alpha1-AT, have been shown to participate in the pathogenesis of human diseases, such as chronic liver-associated diseases[574-576]. The Z mutation resulting from a single G->A transition in codon 342, generates a mutant protein that forms aggregates in the hepatocytes[577]. Liver injury is caused by the retention of this polymerized mutant alpha1-ATZ molecule in the ER of hepatocytes followed by an induction of autophagic response. Removal of the insoluble alpha-1 anti-trypsin by the autophagosome is the mechanism by which the activation of autophagy protects the liver in alpha1-antitrypsin deficiency[578-581]. In earlier studies, liver injury was associated with mitophagy indicating that the ER retention of alpha(1)-ATZ led to involvement of the mitochondria, with specific patterns of mitochondrial dysfunction and mitochondrial injury[582,583].
Genetic studies in mice have shown that deletion of Atg5 led to an increased retention of alpha 1-ATZ[584] and that deficiency of Atg6 and Atg14 in yeasts inhibited alpha1-ATZ degradation[585]. Similarly, the induction of autophagy in mice by rapamycin reduced liver alpha1-ATZ aggregation and liver injury[582,586-589]. These findings have been repeated and verified when enhancement of autophagy[590] with either carbamazepine[591], gene transfer of the autophagy regulator TFEB[592] or an analog of glibenclamide[593] reduced the toxic protein. Recent preclinical studies have also demonstrated that an exogenous bile acid like norursodeoxycholicacid may be clinically useful in this condition[594,595].
Fibrinogen storage disease is a very rare autosomal-dominant ER storage disease presented with hypofibrinogenemia, elevated transaminases, accumulation of fibrinogen aggregates in the ER of hepatocytes and several fold increase of autophagocytic vacuoles. Some patients progress to cirrhosis similar to alpha-1-AT deficiency. A clinical study of eight patients has showed that administration of carbamazepine at low anticonvulsive dosage led to rapid normalization of alanine-aminotransferase indicating a critical role of autophagy in this disease[596].
Wilson disease is an inherited disease of copper metabolism linked to hundreds of mutations in the ATP7B gene[597]. Recent evidence based on studies from hepatocytes of patients and ATP7b deleted mice has shown the presence of an increased number of autophagosomes, indicating the activation of an autophagic response to prevent copper associated cell death[598]. Moreover, inhibition of autophagy accelerated hepatocyte death whereas increased autophagy by either starvation or TFEB overexpression had a cytoprotective effects[598]. Autophagy therefore seems to be a major protective mechanism for hepatocytes in copper accumulation. These findings may lead to the use of autophagy inducers like carbamazepine as a future potential treatment of Wilson’s disease.
Glycogen storage disease type 1a (GSD1a) is an inherited hepatic disease associated with decreased autophagic flux as a consequence of defects in the glucose-6-phosphatase a, that converts glucose-6-phosphate into glucose. These abnormalities lead to glycogen and lipid accumulation in hepatocytes[599]. GSD1a is associated with the down regulation of several components of the autophagy machinery[600].
GSD1a has also been associated with defective sirtuin 1 (SIRT1) signaling leading to impairment of TFEB activity. As in other storage diseases, pharmacological or genetic activation of autophagy reduces the accumulation of glycogen and lipids in cellular and animal models[601].
Autophagy in the liver is implicated in other diseases as well. An important point that should always be remembered is that the liver is the site of almost 80% of body macrophages and therefore innate immunity can be deeply involved in liver and other organ abnormalities through impaired autophagy of Kupffer cells. Sepsis is the main paradigm of this notion.
Infection can lead to a systemic multi-organ inflammatory response. Macrophages, play a critical role as they are the most important cells of the innate immunity. Autophagy induction is protective in sepsis through regulation of macrophage polarization. Negative regulation of macrophage activation inhibits inflammasome activation[602]. Autophagy also interferes with macrophage apoptosis. Uncontrolled autophagy however may lead to autophagy death of macrophages with additional aggravation of inflammation and the so called cytokine storm[603].A current example is possibly the SARS-Cov-2 pandemic[604]. Interestingly, autophagy-deficient macrophages after LPS stimulation over-secrete macrophage migration inhibitory factor and aggravate inflammation[605]. Other mechanisms are also involved including signaling pathways such as NF-κB, mTOR, and PI3K/AKT[603].
Mitophagy and mitochondrial dysfunction seem to be also a fundamental factor in multiple organ failure caused by sepsis[606]. It has been shown that mtDNA liberated from damaged mitochondria, induces a cascade of inflammatory responses[607-609]. Mitophagy therefore is of great importance for the protection against oxidative stress during sepsis. It should be noted however that mitophagy defects in the liver, are not the only cause of organ or cell damage during sepsis[610]. Nonetheless, the liver is the main organ responsible for sepsis-induced damage[611]. Autophagy is an important protective mechanism in septic liver injury. Increased autophagy can play a protective role in liver function in septic conditions where the activation of autophagy is mediated through activating transcription factor 4 (ATF4). ATF4 is inhibited 48 h after LPS-induced acute liver injury and reversed after obeticholic acid treatment[612]. Autophagy inhibitors or AMPK inhibitors administration reduced the protective mitochondrial function in LPS-induced human hepatocyte injury[613,614]. Mitophagy is also involved in apoptosis of CD4+ve T cells which is the main mechanism of immune inhibition during sepsis. Mitofusin 2 (Mfn2) is a mitochondrial outer membrane protein and a negative regulator of autophagy which is increased in sepsis leading to inhibition of autophagy and increase in apoptosis of CD4ve+ T cells[615]. Autophagy defects can affect antigen presentation by T cells leading to immunosuppression as in the case of Atg5 deficiency[616]. The role of autophagy in sepsis has been recently reviewed[617].
Autophagy is also implicated in acetaminophen induced liver disease. There is evidence that increased autophagy is protective against acetaminophen (APAP)-induced liver damage[618,619]. Pathogenetically, APAP was reported to form APAP-protein adducts in hepatocytes of mice and humans[620]. Adducts localized in mitochondria contribute to APAP-induced mitochondrial dysfunction and subsequent oxidant stress[621,622]. Therefore, it is plausible that removal of APAP-adducts will help to ameliorate APAP-induced mitochondrial damage and maintain hepatocyte integrity[41,623,624]. Experimental evidence indicates that autophagy is mostly responsible for the removal of APAP-adducts[625]. Moreover administration of adiponectin was found to attenuate APAP-induced injury activating AMPK mediated autophagy[626]. Activation of autophagy by rapamycin also attenuates APAP-induced liver injury, whereas inhibition of autophagy by chloroquine or deletion of Atg7 in hepatocytes deteriorates liver damage[153,627]. There is also evidence that autophagy is activated after APAP overdose in specific liver zones[53].
Somewhat different results were recently presented. Unc-51-like autophagy activating kinase 1 and 2 (Ulk1/2) are important autophagy initiation regulators. Unexpectedly, Ulk1/2 double knockout mice have normal autophagic activity after fasting, but are exceptionally resistant to APAP-induced liver injury possibly indicating that autophagy-dependent and independent ULK1/2 pathways have opposing effects in APAP-induced liver injury[628].
Reduction of ROS and repression of apoptosis by autophagy is also essential for hepatic regeneration after APAP-induced acute liver failure[520,627]. A very recent report confirmed that increased autophagy by rapamycin protects mice against APAP hepatotoxicity while chloroquine enhanced liver injury. Importantly it was demonstrated that APAP overdose activated PINK1/Parkin-mediated mitophagy and increased the expression of NF-kB and NLRP3 inflammasome signaling. These findings were reversed by rapamycin and augmented by chloroquine indicating the critical role of mitophagy in APAP hepatotoxicity[629].
Interestingly it was reported that infusion of human amniotic mesenchymal stromal cells ameliorated the APAP liver injury through promotion of Kupffer cell M2 polarization and reduction of Kupffer cell autophagy. These results suggest that Kupffer cell autophagy has an opposite effect on APAP hepatotoxicity compared to hepatocytes. This last observation may be useful for future therapeutic exploitation[630].
Acute liver failure (ALF) is a serious syndrome of different etiologies with high mortality[631]. HSCs implication is significant in ALF. Temporarily increased fibrosis in ALF is probably beneficial serving as scaffolding that maintains regenerating hepatocytes and hepatic integrity[437,632,633]. Data from a murine APAP induced ALF model have demonstrated that mortality was significantly increased in HSCs depleted animals[633]. HSCs cannot usually regenerate during ALF due to the submassive necrosis. Autophagy seems to be implicated[634]. The significance of HSCs survival has been verified in a study of patients with HBV induced acute liver failure. ALF was accompanied by fibrosis and HSCs activation and autophagy induction. It was shown for the first time that the High Mobility Group Box 1 (HMGB1) protein is a powerful inducer of autophagy responsible for HSCs survival[635].
As mentioned before, autophagy is crucial for HSCs activation which in turn maintains the liver architecture thus preventing the liver scaffold collapse during ALF.
Nitric oxide induces HSCs apoptosis through generation of ROS[636]. There is evidence however that nitric oxide is also involved in the regulation of autophagy in ALF. Observations in human liver tissue showed an inhibition of autophagy in HSCs while further in vitro experiments demonstrated that nitric oxide inhibited autophagy and increased apoptosis of HSCs. These findings were reproduced by chloroquine and reversed by the autophagy inducer rapamycin. Therefore, nitric oxide impairment of HSCs survival may be a decisive factor for the devastating effects of ALF[637].
An additional clinical and experimental study verified the significance of intact mitophagy in ALF. One of the measurements of oxidative stress is the level of superoxide dismutase (SOD). The serum superoxide dismutase was significantly increased in ALF patients, correlating with the MELD-Na score. SOD levels returned to normal in the remission stage of ALF. In liver tissue from ALF patients and mice models, manganese-dependent SOD was overexpressed and mitophagy in HSCs was inhibited by ROS. Inhibition of mitophagy promoted inflammation in HSCs which was reversed by a mitophagy inducer[454].
Autophagy also protects hepatocytes from acute liver injury, a characteristic of viral hepatitis and acute alcoholic and non-alcoholic steatohepatitis. Mechanisms and cells involved are different as both direct and indirect effects on hepatocytes and macrophages are implicated. Direct effects include autophagy dependent inhibition of caspase 8 in hepatocytes[638], while indirect effects on macrophages involve limitation of NF-kB-mediated inflammation and inflammasome-dependent IL-1b production through p62-dependent mitophagy[462,639]. Reduced macrophage autophagy can induce pro-inflammatory macrophage polarization and increase the immune mediated acute damage in obese mice[131]. The TAM family of RTKs (receptor tyrosine kinases), which is expressed in macrophages, has been reported to alleviate inflammation. AXL is the only member of the TAM family that induces autophagy in macrophages and ameliorates hepatic inflammatory responses inhibiting the NLRP3 inflammasome activation in murine macrophages[640].
The role of Kupffer cells (Kcs) is significant in the pathogenesis of acute liver injury. In a murine model of thioacetamide induced acute liver injury it was shown that hyperglycemia aggravated the liver lesions activating the NLRP3 inflammasome of Kupffer cells via inhibition of AMPK/mTOR-mediated autophagy. Interestingly, AMPK activation or mTOR signaling deletion restored autophagy and subsequently inhibited inflammasome activation in Kupffer cells[641]. Spermine is an anti-oxidative polyamine with autophagy induction properties. In a model of acute liver injury, spermine pre-treatment ameliorated liver injury and intrahepatic inflammation by promoting M2 polarization of Kupffer cells.
Furthermore, spermine increased autophagy in KCs. Deletion of Atg5 in spermine treated KCs greatly increased pro-inflammatory cytokines and reduced the anti-inflammatory cytokine IL-10[642].
LSECs are also involved in acute liver injury. Selective impairment of autophagy in liver endothelial cells increases oxidative stress, thus leading to fibrosis in acute injury[465].
The central role of autophagy in ischemia/reperfusion injury (I/R) injury has been verified by the fact that pharmacological or genetic stimulation of autophagy ameliorate the liver reperfusion injury[643-645].
I/R impairs hepatocellular autophagy[646] through I/R-induced ATP depletion leading to energy shortage and malfunction of the autophagic machinery. Moreover Ca2+ overloading during I/R results in calpain overproduction and ultimate loss of key autophagy proteins like Atg7. Interestingly the autophagy suppressor chloroquine attenuated liver injury when administered in early phases of I/R but aggravated the lesions, as expected, when given in late phases[647].
Ammonia is an important mediator of hepatic encephalopathy. Increased ammonia levels rapidly induce an autophagic response that preferentially targets mitophagy[648-650]. Ammonia induced autophagy may in fact be a protective mechanism against encephalopathy as suggested by a recent report. Deletion of Atg7 or loss of functional TFEB deteriorated ammonia detoxification in mice. By contrast activation of liver autophagy either by rapamycin administration or genetic TFEB expression reduced ammonia levels in acquired hyperammonaemia[651].
The role of autophagy in autoimmune hepatitis (AIH) has not been adequately studied. It is suggested that autophagy is implicated in AIH through its involvement in antigen processing and presentation to T cells[652] and its well proven role in liver fibrosis[653], but the exact pathways have not been delineated.
Concanavalin A-induced hepatitis is an extensively used model for immune-mediated liver injury. Comparative proteomic results in this model have shown that the activation of immune system resulted in hepatitis with deregulation of autophagy as indicated by an increase in p62 and LC3B. Arctigenin is a biologically active lignan with antioxidant and anti-inflammatory properties. Pretreatment with arctigenin alleviated autophagy as well as apoptosis verifying that immunity and autophagy are interconnected in AIH pathogenesis[654].
A group of researchers recently used the same model of concanavalin (conA) induced experimental hepatitis to clarify the role of autophagy in AIH. Methyl prednisolone (MP) treatment significantly decreased inflammation in the liver and activated the Akt/mTOR pathway to inhibit hepatocyte apoptosis and autophagy. Reduced numbers of autophagosomes were present in the MP treated group compared to the conA group. It was further shown that MP attenuated the mitochondria-mediated autophagy and apoptosis[655]. In a second report on the same experimental model, accumulation of mature conventional dendritic cells (cDCs) was observed in the liver. In vitro, ConA treatment induced the expression of autophagy proteins and the formation of autophagosomes in dendritic cells. A further blockade of autophagy flux inhibited the maturation of DCs and the proliferation and differentiation of CD4+ T cells when ConA-induced DCs were co-cultured with CD4+ T cells. Taken together these studies elegantly showed that autophagy is critically implicated in AIH and aberrant autophagy and defective maturation of cDCs are involved in AIH immunopathogenesis[656].
A recent clinical study using immunohistochemistry in liver biopsy samples from chronic HCV and AIH patients confirmed the central role of autophagy in AIH. Activated but impaired autophagy and less efficient elimination of damaged mitochondria were demonstrated in AIH as compared with HCV. Increased p62 levels significantly correlated with necroinflammation in AIH[657].
The mechanisms of liver damage in cholestasis are incompletely understood. Autophagy and protein degradation were shown to be impaired in cholestasis induced in bile duct ligated mice[658-660]. Moreover, defective autophagy after chloroquine inhibition or deletion of Atg7 and Atg5 led to increased cholestatic liver injury[661,662].
Accumulated toxic bile acids lead to ER stress, mitochondrial dysfunction with increased oxidative stress, inflammasome activation and apoptosis leading to liver fibrosis[663]. These events should in fact activate autophagy in cholestasis but instead, at least in mice, it appears that autophagy is inhibited in cholestasis[664,665]. Bile acids can inhibit autophagy in mice either via the farnesoid X receptor (FXR) during the feeding-fasting cycle[114,115] or independently of FXR[666]. How autophagy is affected in human cholestasis is under investigation.
In human disease autophagy was initially associated with the pathogenesis of primary biliary cholangitis (PBC)[667-669]. As mentioned before autophagy is also involved in the processing and presentation of various antigens. It is only logical therefore that an interesting hypothesis implicating deregulated cholangiocyte autophagy connected to cholangiocyte senescence has been proposed to explain not only the pathogenesis of PBC but of the other fibrosing cholangiopathies including primary sclerosing cholangitis (PSC) and biliary atresia as well[670].
An upregulation of autophagy was reported along with senescence in PBC[668,671]. LC3B and p62 proteins were accumulated in damaged bile ductular cells in association with senescence markers[68,125] suggesting that autophagy could induce and facilitate cholangiocyte senescence[664,665,671-674]. Mitophagy may be specifically involved in PBC as granular expression of the mitochondrial protein PDC-E2 was co-localized with LC3[667].
Autophagy has also been implicated in the treatment of PBC. Ursodeoxycholic acid (UDCA) is still the first line treatment of PBC while obeticholic acid (OCA) is a second-line treatment[675-677]. Hydrophobic bile acids, such as glycochenodeoxycholic acid impair autophagy in vitro and induce abnormal expression of mitochondrial antigens and cellular senescence in cholangiocytes, possibly through induction of ER stress. Pretreatment with UDCA reduced ER stress and partially restored deregulated autophagy and cellular senescence[678]. It is not clear how UDCA stimulates autophagy. UDCA has been reported to be an FXR antagonist[679] but this may not be the explanation[680]. On the contrary, OCA is a semi-synthetic FXR agonist with anti-cholestatic functions including the suppression of endogenous bile acid synthesis and interference with hepatocellular bile acid transporter systems[681]. OCA impairs autophagic flux in vitro and also in vivo. A favorable effect of treatment with OCA in a cholestatic disease like PBC would be incompatible with data, indicating that cholestasis progresses when autophagy is blocked[661,662]. However, the other potent, anti-cholestatic properties of OCA can overcome the negative effects of reduced autophagy.
A recent paper offers an interesting explanation. Autophagy seems to be also impaired in human cholestatic conditions where accumulated bile acids induce Rubicon in an FXR-dependent fashion. Rubicon induction suppresses autophagosome-lysosome fusion and inhibits proper autophagolytic breakdown. Rubicon was also induced after treatment with the FXR agonist OCA. Genetic inhibition of Rubicon reversed the impairment of autophagic flux. In contrast, UDCA reduced Rubicon levels, enhanced autophagic flux and autophagolysosome formation independently of FXR[680].
An overview of autophagy abnormalities is presented in Table 1.
| Disease | Abnormalities of autophagy | Results | Ref. |
| Obesity | ↓Autophagy; Hepatocytes: ↓Mitophagy, ↓Lipophagy; HSCs: ↓Autophagy | ↑ER stress, →↑Lipids, ↑Insuline resistance, → Anti-fibrotic | Liu et al[203], Lavallard et al[204], Gual et al[205], Tremblay et al[206] |
| NAFLD | ↓Lipophagy; ↓CMA | Lipotoxicity, ↑Lipogenic enzymes | Madrigal-Matute et al[30], Zhou et al[234], Niso-Santano et al[235], Singh et al[236] |
| NASH | Hepatocytes: ↓Autophagy, ↓Mitophagy; Kupffer cells: ↓Autophagy; LSECs: ↓Autophagy | ↑Mallory-Denk bodies, ↑Inflammasome activation; ↑Cathepsins B,D, ↑M1 polarization, ↓M2 polarization; ↑Inflammation, fibrosis | Xu et al[272], Noureddin et al[277], Zhang et al[285], Dey et al[287] |
| Alcoholic liver disease | Acute ETOH administration: ↑Autophagy, ↑Mitophagy, ↑Lipophagy, ↑Proteophagy; Chronic ETOH administration: ↑ Autophagy (low dose), ↓Autophagy (high dose); Kupffer cells: ↓Autophagy, ↑Autophagy; HSCs: ↓Autophagy, ↑Autophagy | Protection, protection, protection, →Clearance of Mallory-Denk bodies; →Protection, →Mitochondrial damage, Cell death; Liver damage, protection; Reduced fibrosis, increased fibrosis | Chao et al[308], Komatsu et al[311], Yan et al[314], Harada et al[318] |
| HBV | ↑Autophagy, ↓Lysosomal acidification, ↑Mitophagy | ↑Virus replication, ↓HBV degradation | Li et al[356], Tang et al[357], Luo et al[372], Wang et al[383] |
| HCV | ↑Autophagy, ↓Lipophagy, ↑Mitophagy; ↑CMA | ↑Virus replication, steatosis, ↑Virus replication, ↓Apoptosis, persitent infection, ↑Virus replication | Ferraris et al[387], Paul et al[395], Jassey et al[404], Ren et al[406] |
| Fibrosis-Cirrhosis | Hepatocytes: ↓Autophagy, ↓Lipophagy; Kupffer cells: ↓Mitophagy, or, ↑↑Mitophagy; HSCs: ↓Mitophagy, ↓Lipophagy, or, ↑Lipophagy, ↑Mitophagy; LSECs: ↑↓Autophagy; Ductular reaction: ↑Autophagy | ↑Fibrosis, ↑Lipotoxicity, ↓TGFb, ↓Fibrosis; ↑TGFb, ↑Fibrosis; Pro-inflammatory anti-fibrotic: →Pro-fibrotic, →Pro-fibrotic, ↑Fibrosis, ↑Fibrosis | Zhang et al[437], Singh et al[438], Li et al[448], Sun et al[463] |
| HCC, “Double edge sword” | Induction stage: ↑CMA, ↑Autophagy; Late stages: ↑Autophagy, or, ↓Autophagy, ↑Mitophagy, ↑Lipophagy | Anti-oncogenic: ↓YAP1, ↓proliferation, ↑Apoptosis→Anti-oncogenic, ↓Tumor suppressors; ↑Tumor progression, ↓↑Progression↑↓Progression | Wang et al[558], Zhao et al[559], Prieto-Domínguez et al[560]; Niture et al[544], Yang et al[547]; Lin et al[549], Chen et al[550], Chen et al[551] |
| Cholangiocarcinoma | ↑Autophagy | ↑Tumor progression | Marciniak et al[580], Teckman et al[581] |
| A1 antitrypsin deficiency | ↓Autophagy | Yamamura et al[590], Pastore et al[592] | |
| Fibrinogen storage disease | ↓Autophagy | Hu et al[609] | |
| Wilson’S disease | ↓Autophagy | Oami et al[611] | |
| Glycogen storage disease | ↓Autophagy | Xing et al[613] | |
| Sepsis | Kupffer cells: ↑Autophagy, ↑↑Autophagy, ↓Mitophagy | M2 polarization, ↓Inflammasome activation; Kupffer cell apoptosis→Cytokine storm, ↓Apoptosis of CD4+ve T cells | Ying et al[615], Neumann et al[616], Sun et al[628], Shan et al[629] |
| Acetaminophene liver damage | ↓Autophagy, ↓Mitophagy, ↑Kupffer cell autophagy | ↑APAP-Protein adducts | Sydor et al[618], Kim et al[643], Biel et al[644] |
| Acute liver failure | ↑Autophagy, ↓Autophagy, ↓HSCs Mitophagy | HMGB1→HSCs activation (protective); ↑NO,ROS→↓HSCs→Devastation | Cheong et al[649], Sridhar et al[652] |
| Ischemia/reperfusion injury | ↓Autophagy | Kwak et al[658], Huang et al[659] | |
| Hepatic encephalopathy | ↑Autophagy (NH4) | Protection | Woolbright et al[663], Manley et al[666] |
| Autoimmune hepatitis | ↑Autophagy, ↓ Mitophagy | Defective maturation of dendritic cells | Sasaki et al[671], Sasaki et al[672], Young et al[673] |
| Biliary disease (experimental) | ↓Autophagy | Possibly through increased bile acids | Sasaki et al[665], European Association for the Study of the Liver[675], Lindor et al[676], Panzitt et al[680] |
| Primary biliay cholangitis | Deregulated autophagy | Cholangiocyte senescence | Van de Graaf et al[669], Sasaki et al[665], Sasaki et al[674] |
Autophagy is an important process through which intracellular parts are degraded in the lysosomes. It is a fine example of effective cellular recycling mechanism, connecting cellular quality control with energy saves. There are three types of autophagy with various pathways of delivery to the lysosomes: Macroautophagy (which is further divided into non selective autophagy and selective macroautophagy targeting special organelles or specific compounds for degradation), microautophagy and chaperon-mediated autophagy. Autophagy is related to major physiologic processes as cell death, inflammation and immunity. It is increasingly recognized that it is implicated in almost every aspect of liver diseases, and this can be the basis for future pathophysiologically based and targeted management.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pan Q, Tavernarakis N S-Editor: Gao CC L-Editor: A P-Editor: Wang LL
| 1. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651-662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1792] [Cited by in F6Publishing: 1842] [Article Influence: 167.5] [Reference Citation Analysis (0)] |
| 2. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1024] [Cited by in F6Publishing: 1703] [Article Influence: 340.6] [Reference Citation Analysis (0)] |
| 3. | Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem. 1981;256:7652-7658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 95] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA. 1983;80:2179-2183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 132] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Ktistakis NT. In praise of M. Anselmier who first used the term "autophagie" in 1859. Autophagy. 2017;13:2015-2017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | De Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604-617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3002] [Cited by in F6Publishing: 3238] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 7. | Novikoff AB, Beaufay H, De Duve C. Electron microscopy of lysosomerich fractions from rat liver. J Biophys Biochem Cytol. 1956;2:179-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 156] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Appelmans F, Wattiaux R, De Duve C. Tissue fractionation studies. 5. The association of acid phosphatase with a special class of cytoplasmic granules in rat liver. Biochem J. 1955;59:438-445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 450] [Cited by in F6Publishing: 536] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 9. | Essner E, Novikoff AB. Human hepatocellular pigments and lysosomes. J Ultrastruct Res. 1960;3:374-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 272] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 10. | De Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 266] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 11. | Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687-733. [PubMed] [Cited in This Article: ] |
| 12. | Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 887] [Cited by in F6Publishing: 901] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 13. | Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1305] [Cited by in F6Publishing: 1334] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 14. | Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 285] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 15. | Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 880] [Cited by in F6Publishing: 922] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 16. | Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B, Mizushima N, Rubinsztein DC, Thumm M, Tooze SA. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:438-448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 17. | Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, Finkbeiner S, Fueyo-Margareto J, Gewirtz D, Jäättelä M, Kroemer G, Levine B, Melia TJ, Mizushima N, Rubinsztein DC, Simonsen A, Thorburn A, Thumm M, Tooze SA. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy. 2011;7:1273-1294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 18. | Sheng R, Qin ZH. History and Current Status of Autophagy Research. Adv Exp Med Biol. 2019;1206:3-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Harnett MM, Pineda MA, Latré de Laté P, Eason RJ, Besteiro S, Harnett W, Langsley G. From Christian de Duve to Yoshinori Ohsumi: More to autophagy than just dining at home. Biomed J. 2017;40:9-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 20. | Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 625] [Cited by in F6Publishing: 738] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 21. | Levine B, Klionsky DJ. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker's yeast fuel advances in biomedical research. Proc Natl Acad Sci USA. 2017;114:201-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 113] [Article Influence: 14.1] [Reference Citation Analysis (1)] |
| 22. | Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521-527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 468] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 23. | Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831-835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 405] [Cited by in F6Publishing: 414] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 24. | Li Y, Ding WX. Adipose tissue autophagy and homeostasis in alcohol-induced liver injury. Liver Res. 2017;1:54-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 911] [Cited by in F6Publishing: 940] [Article Influence: 134.3] [Reference Citation Analysis (0)] |
| 26. | Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1271] [Cited by in F6Publishing: 1802] [Article Influence: 360.4] [Reference Citation Analysis (0)] |
| 27. | Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201-2214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 342] [Cited by in F6Publishing: 363] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 28. | Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion. J Cell Sci. 2017;130:1209-1216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 296] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 29. | Sarkar C, Zhao Z, Aungst S, Sabirzhanov B, Faden AI, Lipinski MM. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy. 2014;10:2208-2222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 236] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 30. | Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology. 2016;150:328-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 228] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 31. | Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101-1111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1719] [Cited by in F6Publishing: 1892] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 32. | Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 812] [Cited by in F6Publishing: 888] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 33. | Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3327] [Cited by in F6Publishing: 3662] [Article Influence: 261.6] [Reference Citation Analysis (0)] |
| 34. | Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Bánhegyi G, Bartholomew CR, Bassham DC, Bast RC Jr, Batoko H, Bay BH, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronjé MJ, Cuervo AM, Cullen JJ, Czaja MJ, D'Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farré JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, González-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He C, He YW, Hébert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Høyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jiménez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhász G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, László L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, López-Otín C, Lossi L, Lotze MT, Lőw P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Mariño G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Meléndez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Müller S, Muller S, Münger K, Münz C, Murphy LO, Murphy ME, Musarò A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nürnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Pöggeler S, Poirot M, Poletti A, Poüs C, Pozuelo-Rubio M, Prætorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reiners JJ Jr, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodríguez de Córdoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB 3rd, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sánchez-Alcázar JA, Sánchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schätzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schüller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Strålfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takács-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tönges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ 3rd, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2782] [Cited by in F6Publishing: 2768] [Article Influence: 230.7] [Reference Citation Analysis (0)] |
| 35. | Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603-614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2315] [Cited by in F6Publishing: 2551] [Article Influence: 134.3] [Reference Citation Analysis (0)] |
| 36. | Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1783] [Cited by in F6Publishing: 2030] [Article Influence: 119.4] [Reference Citation Analysis (0)] |
| 37. | Islam MA, Sooro MA, Zhang P. Autophagic Regulation of p62 is Critical for Cancer Therapy. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 38. | Zhang NP, Liu XJ, Xie L, Shen XZ, Wu J. Impaired mitophagy triggers NLRP3 inflammasome activation during the progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis. Lab Invest. 2019;99:749-763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 39. | Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2082] [Cited by in F6Publishing: 2255] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 40. | Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149-1163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1613] [Cited by in F6Publishing: 1681] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 41. | Czaja MJ, Ding WX, Donohue TM Jr, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, Perlmutter DH, Randall G, Ray RB, Tsung A, Yin XM. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 351] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 42. | Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773-1785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 43. | Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jäättelä M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Münz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811-1836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1182] [Cited by in F6Publishing: 1130] [Article Influence: 161.4] [Reference Citation Analysis (0)] |
| 44. | Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4208] [Cited by in F6Publishing: 5098] [Article Influence: 392.2] [Reference Citation Analysis (0)] |
| 45. | Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456-461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1966] [Cited by in F6Publishing: 1930] [Article Influence: 148.5] [Reference Citation Analysis (0)] |
| 46. | Angarola B, Ferguson SM. Coordination of Rheb lysosomal membrane interactions with mTORC1 activation. F1000Res. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 47. | Raben N, Puertollano R. TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Annu Rev Cell Dev Biol. 2016;32:255-278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 283] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 48. | Settembre C, Ballabio A. TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy. 2011;7:1379-1381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 49. | Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429-1433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2278] [Cited by in F6Publishing: 2343] [Article Influence: 180.2] [Reference Citation Analysis (0)] |
| 50. | Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1070] [Cited by in F6Publishing: 1151] [Article Influence: 104.6] [Reference Citation Analysis (0)] |
| 51. | Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903-914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 756] [Cited by in F6Publishing: 932] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 52. | Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 773] [Cited by in F6Publishing: 961] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 53. | Chao X, Wang H, Jaeschke H, Ding WX. Role and mechanisms of autophagy in acetaminophen-induced liver injury. Liver Int. 2018;38:1363-1374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 54. | Santambrogio L, Cuervo AM. Chasing the elusive mammalian microautophagy. Autophagy. 2011;7:652-654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 55. | Olsvik HL, Svenning S, Abudu YP, Brech A, Stenmark H, Johansen T, Mejlvang J. Endosomal microautophagy is an integrated part of the autophagic response to amino acid starvation. Autophagy. 2019;15:182-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 56. | Ke PY. Diverse Functions of Autophagy in Liver Physiology and Liver Diseases. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 57. | Oku M, Sakai Y. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. Bioessays. 2018;40:e1800008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 58. | Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365-381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 547] [Cited by in F6Publishing: 787] [Article Influence: 157.4] [Reference Citation Analysis (0)] |
| 59. | Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J Biol Chem. 2018;293:5414-5424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 242] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 60. | Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 61. | Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200-C1208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 253] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 62. | Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829-4840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 452] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 63. | Dohi E, Tanaka S, Seki T, Miyagi T, Hide I, Takahashi T, Matsumoto M, Sakai N. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem Int. 2012;60:431-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 64. | Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864-25870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 104] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 65. | Ferreira JV, Fôfo H, Bejarano E, Bento CF, Ramalho JS, Girão H, Pereira P. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349-1366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 66. | Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 611] [Cited by in F6Publishing: 580] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 67. | Farré JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17:537-552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 290] [Cited by in F6Publishing: 274] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 68. | Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 543] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 69. | Dunn WA Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 202] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 70. | Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 728] [Cited by in F6Publishing: 1190] [Article Influence: 119.0] [Reference Citation Analysis (0)] |
| 71. | Nakatogawa H, Mochida K. Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy. 2015;11:2377-2378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 72. | Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2517] [Cited by in F6Publishing: 2887] [Article Influence: 192.5] [Reference Citation Analysis (0)] |
| 73. | Lee S, Kim JS. Mitophagy: therapeutic potentials for liver disease and beyond. Toxicol Res. 2014;30:243-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 74. | Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, Katona I, Qualmann B, Weis J, Reggiori F, Kurth I, Hübner CA, Dikic I. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 548] [Cited by in F6Publishing: 649] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 75. | Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 464] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 76. | Ellgaard L, Sevier CS, Bulleid NJ. How Are Proteins Reduced in the Endoplasmic Reticulum? Trends Biochem Sci. 2018;43:32-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 77. | Grumati P, Dikic I, Stolz A. ER-phagy at a glance. J Cell Sci. 2018;131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 78. | Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 554] [Cited by in F6Publishing: 741] [Article Influence: 123.5] [Reference Citation Analysis (0)] |
| 79. | Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, Wilkinson S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell 2018; 44: 217-232. e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 296] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 80. | Ding WX, Manley S, Ni HM. The emerging role of autophagy in alcoholic liver disease. Exp Biol Med (Maywood). 2011;236:546-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 81. | Wang L, Khambu B, Zhang H, Yin XM. Autophagy in alcoholic liver disease, self-eating triggered by drinking. Clin Res Hepatol Gastroenterol. 2015;39 Suppl 1:S2-S6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 82. | Zubiete-Franco I, García-Rodríguez JL, Martínez-Uña M, Martínez-Lopez N, Woodhoo A, Juan VG, Beraza N, Lage-Medina S, Andrade F, Fernandez ML, Aldámiz-Echevarría L, Fernández-Ramos D, Falcon-Perez JM, Lopitz-Otsoa F, Fernandez-Tussy P, Barbier-Torres L, Luka Z, Wagner C, García-Monzón C, Lu SC, Aspichueta P, Mato JM, Martínez-Chantar ML, Varela-Rey M. Methionine and S-adenosylmethionine levels are critical regulators of PP2A activity modulating lipophagy during steatosis. J Hepatol. 2016;64:409-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 83. | Xiong J, Wang K, He J, Zhang G, Zhang D, Chen F. TFE3 Alleviates Hepatic Steatosis through Autophagy-Induced Lipophagy and PGC1α-Mediated Fatty Acid β-Oxidation. Int J Mol Sci. 2016;17:387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 84. | Chen K, Yuan R, Zhang Y, Geng S, Li L. Tollip Deficiency Alters Atherosclerosis and Steatosis by Disrupting Lipophagy. J Am Heart Assoc. 2017;6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 85. | Hung TM, Yuan RH, Huang WP, Chen YH, Lin YC, Lin CW, Lai HS, Lee PH. Increased Autophagy Markers Are Associated with Ductular Reaction during the Development of Cirrhosis. Am J Pathol. 2015;185:2454-2467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 86. | Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, Ou JH. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025-1034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 87. | Zhang Z, Zhao S, Yao Z, Wang L, Shao J, Chen A, Zhang F, Zheng S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol. 2017;11:322-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 88. | Martinez-Lopez N, Singh R. Autophagy and Lipid Droplets in the Liver. Annu Rev Nutr. 2015;35:215-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 227] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 89. | Schulze RJ, Sathyanarayan A, Mashek DG. Breaking fat: The regulation and mechanisms of lipophagy. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1178-1187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 151] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 90. | Zhang Z, Yao Z, Chen Y, Qian L, Jiang S, Zhou J, Shao J, Chen A, Zhang F, Zheng S. Lipophagy and liver disease: New perspectives to better understanding and therapy. Biomed Pharmacother. 2018;97:339-348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 91. | Kounakis K, Chaniotakis M, Markaki M, Tavernarakis N. Emerging Roles of Lipophagy in Health and Disease. Front Cell Dev Biol. 2019;7:185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 92. | Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879-27890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 441] [Cited by in F6Publishing: 472] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 93. | Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 710] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 94. | Menzies RA, Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J Biol Chem. 1971;246:2425-2429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 255] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 95. | Pfeifer U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver. A morphometric approach to the kinetics of intracellular degradation by autophagy. J Cell Biol. 1978;78:152-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 230] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 96. | Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 839] [Cited by in F6Publishing: 929] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 97. | Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245-253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1138] [Cited by in F6Publishing: 1202] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 98. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4642] [Cited by in F6Publishing: 5103] [Article Influence: 318.9] [Reference Citation Analysis (0)] |
| 99. | Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2130] [Cited by in F6Publishing: 2363] [Article Influence: 181.8] [Reference Citation Analysis (0)] |
| 100. | Zhao L, Li H, Wang Y, Zheng A, Cao L, Liu J. Autophagy Deficiency Leads to Impaired Antioxidant Defense via p62-FOXO1/3 Axis. Oxid Med Cell Longev. 2019;2019:2526314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 101. | Lemasters JJ. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol. 2014;2:749-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 102. | Flores-Toro JA, Go KL, Leeuwenburgh C, Kim JS. Autophagy in the liver: cell's cannibalism and beyond. Arch Pharm Res. 2016;39:1050-1061. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 103. | Ke PY. Mitophagy in the Pathogenesis of Liver Diseases. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 104. | Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 587] [Cited by in F6Publishing: 615] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 105. | Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, Hu YC. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 176] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 106. | Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473:1268-1275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 107. | Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605-2612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 108. | Xiong H, Ni Z, He J, Jiang S, Li X, He J, Gong W, Zheng L, Chen S, Li B, Zhang N, Lyu X, Huang G, Chen B, Zhang Y, He F. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528-3540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 284] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 109. | Islam Khan MZ, Tam SY, Law HKW. Autophagy-Modulating Long Non-coding RNAs (LncRNAs) and Their Molecular Events in Cancer. Front Genet. 2018;9:750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 110. | Horos R, Büscher M, Kleinendorst R, Alleaume AM, Tarafder AK, Schwarzl T, Dziuba D, Tischer C, Zielonka EM, Adak A, Castello A, Huber W, Sachse C, Hentze MW. The Small Non-coding Vault RNA1-1 Acts as a Riboregulator of Autophagy. Cell 2019; 176: 1054-1067. e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 111. | Zhao Y, Wang Z, Zhang W, Zhang L. MicroRNAs play an essential role in autophagy regulation in various disease phenotypes. Biofactors. 2019;45:844-856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 112. | Khambu B, Huda N, Chen X, Antoine DJ, Li Y, Dai G, Köhler UA, Zong WX, Waguri S, Werner S, Oury TD, Dong Z, Yin XM. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J Clin Invest. 2018;128:2419-2435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 113. | Lee YA, Noon LA, Akat KM, Ybanez MD, Lee TF, Berres ML, Fujiwara N, Goossens N, Chou HI, Parvin-Nejad FP, Khambu B, Kramer EGM, Gordon R, Pfleger C, Germain D, John GR, Campbell KN, Yue Z, Yin XM, Cuervo AM, Czaja MJ, Fiel MI, Hoshida Y, Friedman SL. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat Commun. 2018;9:4962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 114. | Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 402] [Cited by in F6Publishing: 386] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 115. | Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 322] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 116. | Kim H, Williams D, Qiu Y, Song Z, Yang Z, Kimler V, Goldberg A, Zhang R, Yang Z, Chen X, Wang L, Fang D, Lin JD, Zhang K. Regulation of hepatic autophagy by stress-sensing transcription factor CREBH. FASEB J. 2019;33:7896-7914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 117. | Xu J, Camfield R, Gorski SM. The interplay between exosomes and autophagy - partners in crime. J Cell Sci. 2018;131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 217] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 118. | Zheng J, Tan J, Miao YY, Zhang Q. Extracellular vesicles degradation pathway based autophagy lysosome pathway. Am J Transl Res. 2019;11:1170-1183. [PubMed] [Cited in This Article: ] |
| 119. | Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1053] [Cited by in F6Publishing: 1164] [Article Influence: 83.1] [Reference Citation Analysis (0)] |
| 120. | Chen Y, Yu L. Recent progress in autophagic lysosome reformation. Traffic. 2017;18:358-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 121. | Chen Y, Yu L. Development of Research into Autophagic Lysosome Reformation. Mol Cells. 2018;41:45-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 22] [Reference Citation Analysis (0)] |
| 122. | Buratta S, Tancini B, Sagini K, Delo F, Chiaradia E, Urbanelli L, Emiliani C. Lysosomal Exocytosis, Exosome Release and Secretory Autophagy: The Autophagic- and Endo-Lysosomal Systems Go Extracellular. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 212] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 123. | Galluzzi L, Green DR. Autophagy-Independent Functions of the Autophagy Machinery. Cell. 2019;177:1682-1699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 339] [Cited by in F6Publishing: 571] [Article Influence: 142.8] [Reference Citation Analysis (0)] |
| 124. | Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 328] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 125. | Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925-935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 182] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 126. | Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2268] [Cited by in F6Publishing: 2460] [Article Influence: 189.2] [Reference Citation Analysis (1)] |
| 127. | Mihalache CC, Simon HU. Autophagy regulation in macrophages and neutrophils. Exp Cell Res. 2012;318:1187-1192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 128. | Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 943] [Cited by in F6Publishing: 1050] [Article Influence: 87.5] [Reference Citation Analysis (0)] |
| 129. | Puleston DJ, Simon AK. Autophagy in the immune system. Immunology. 2014;141:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 130. | Chen P, Cescon M, Bonaldo P. Autophagy-mediated regulation of macrophages and its applications for cancer. Autophagy. 2014;10:192-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 131. | Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11:271-284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 329] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 132. | Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 424] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 133. | Heckmann BL, Boada-Romero E, Cunha LD, Magne J, Green DR. LC3-Associated Phagocytosis and Inflammation. J Mol Biol. 2017;429:3561-3576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 181] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 134. | Martinez J. LAP it up, fuzz ball: a short history of LC3-associated phagocytosis. Curr Opin Immunol. 2018;55:54-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 135. | Münz C. Non-canonical Functions of Macroautophagy Proteins During Endocytosis by Myeloid Antigen Presenting Cells. Front Immunol. 2018;9:2765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 136. | Schneider JL, Cuervo AM. Liver autophagy: much more than just taking out the trash. Nat Rev Gastroenterol Hepatol. 2014;11:187-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 137. | Cuervo AM, Macian F. Autophagy and the immune function in aging. Curr Opin Immunol. 2014;29:97-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 138. | Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753-766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1625] [Cited by in F6Publishing: 1697] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 139. | Czaja MJ. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2016;61:1304-1313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 138] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 140. | Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, Wu C, Vogel P, Neale G, Green DR, Chi H. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. 2016;17:277-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 329] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 141. | Peral de Castro C, Jones SA, Ní Cheallaigh C, Hearnden CA, Williams L, Winter J, Lavelle EC, Mills KH, Harris J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012;189:4144-4153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 125] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 142. | Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2401] [Cited by in F6Publishing: 2632] [Article Influence: 188.0] [Reference Citation Analysis (0)] |
| 143. | Amelio I, Melino G, Knight RA. Cell death pathology: cross-talk with autophagy and its clinical implications. Biochem Biophys Res Commun. 2011;414:277-281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 144. | Yonekawa T, Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55:105-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 148] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 145. | Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025-1040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1271] [Cited by in F6Publishing: 1297] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 146. | Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2430] [Cited by in F6Publishing: 2716] [Article Influence: 159.8] [Reference Citation Analysis (0)] |
| 147. | Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966-975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 897] [Cited by in F6Publishing: 914] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 148. | Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta. 2013;1833:3448-3459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 844] [Cited by in F6Publishing: 959] [Article Influence: 87.2] [Reference Citation Analysis (0)] |
| 149. | Wang K. Autophagy and apoptosis in liver injury. Cell Cycle. 2015;14:1631-1642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 272] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 150. | Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol. 2005;6:505-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 744] [Cited by in F6Publishing: 760] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 151. | Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679-2688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1271] [Cited by in F6Publishing: 1310] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 152. | Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1403] [Cited by in F6Publishing: 1632] [Article Influence: 163.2] [Reference Citation Analysis (0)] |
| 153. | Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 334] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 154. | Lockshin RA, Zakeri Z. Cell death in health and disease. J Cell Mol Med. 2007;11:1214-1224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 155. | Strozyk E, Kulms D. The role of AKT/mTOR pathway in stress response to UV-irradiation: implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int J Mol Sci. 2013;14:15260-15285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 156. | Liang Q, Xiao Y, Liu K, Zhong C, Zeng M, Xiao F. Cr(VI)-Induced Autophagy Protects L-02 Hepatocytes from Apoptosis Through the ROS-AKT-mTOR Pathway. Cell Physiol Biochem. 2018;51:1863-1878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 157. | Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 548] [Article Influence: 109.6] [Reference Citation Analysis (0)] |
| 158. | Liu J, Kuang F, Kroemer G, Klionsky DJ, Kang R, Tang D. Autophagy-Dependent Ferroptosis: Machinery and Regulation. Cell Chem Biol. 2020;27:420-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 431] [Article Influence: 107.8] [Reference Citation Analysis (0)] |
| 159. | Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021-1032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 612] [Cited by in F6Publishing: 1089] [Article Influence: 136.1] [Reference Citation Analysis (2)] |
| 160. | Hou W, Xie Y, Song X, Sun X, Lotze MT, Zeh HJ 3rd, Kang R, Tang D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425-1428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1171] [Cited by in F6Publishing: 1336] [Article Influence: 167.0] [Reference Citation Analysis (0)] |
| 161. | Kang R, Tang D. Autophagy and Ferroptosis - What's the Connection? Curr Pathobiol Rep. 2017;5:153-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 162. | Bai Y, Meng L, Han L, Jia Y, Zhao Y, Gao H, Kang R, Wang X, Tang D, Dai E. Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun. 2019;508:997-1003. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 284] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 163. | Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677-687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1797] [Cited by in F6Publishing: 2266] [Article Influence: 251.8] [Reference Citation Analysis (0)] |
| 164. | Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1529] [Cited by in F6Publishing: 2187] [Article Influence: 273.4] [Reference Citation Analysis (0)] |
| 165. | Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol. 2015;16:1014-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 348] [Cited by in F6Publishing: 366] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 166. | de Lavera I, Pavon AD, Paz MV, Oropesa-Avila M, de la Mata M, Alcocer-Gomez E, Garrido-Maraver J, Cotan D, Alvarez-Cordoba M, Sanchez-Alcazar JA. The Connections Among Autophagy, Inflammasome and Mitochondria. Curr Drug Targets. 2017;18:1030-1038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 167. | Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253-1257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 968] [Cited by in F6Publishing: 1032] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 168. | Yordy B, Iwasaki A. Autophagy in the control and pathogenesis of viral infection. Curr Opin Virol. 2011;1:196-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 169. | Oh JE, Lee HK. Autophagy as an innate immune modulator. Immune Netw. 2013;13:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 170. | Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821-832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3880] [Cited by in F6Publishing: 4341] [Article Influence: 310.1] [Reference Citation Analysis (0)] |
| 171. | Wang Z, Zhang S, Xiao Y, Zhang W, Wu S, Qin T, Yue Y, Qian W, Li L. NLRP3 Inflammasome and Inflammatory Diseases. Oxid Med Cell Longev. 2020;2020:4063562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 131] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 172. | Thorburn A. Autophagy and its effects: making sense of double-edged swords. PLoS Biol. 2014;12:e1001967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 173. | White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1226] [Cited by in F6Publishing: 1318] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 174. | Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913-1930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 457] [Cited by in F6Publishing: 594] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 175. | Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1278] [Cited by in F6Publishing: 1452] [Article Influence: 132.0] [Reference Citation Analysis (0)] |
| 176. | Codogno P, Meijer AJ. Autophagy in the liver. J Hepatol. 2013;59:389-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 177. | Gual P, Gilgenkrantz H, Lotersztajn S. Autophagy in chronic liver diseases: the two faces of Janus. Am J Physiol Cell Physiol. 2017;312:C263-C273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 178. | Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 358] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 179. | Weiskirchen R, Tacke F. Relevance of Autophagy in Parenchymal and Non-Parenchymal Liver Cells for Health and Disease. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 180. | Allaire M, Rautou PE, Codogno P, Lotersztajn S. Autophagy in liver diseases: Time for translation? J Hepatol. 2019;70:985-998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 258] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 181. | Yan S, Huda N, Khambu B, Yin XM. Relevance of autophagy to fatty liver diseases and potential therapeutic applications. Amino Acids. 2017;49:1965-1979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 182. | Khambu B, Yan S, Huda N, Liu G, Yin XM. Autophagy in non-alcoholic fatty liver disease and alcoholic liver disease. Liver Res. 2018;2:112-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 183. | Khambu B, Yan S, Huda N, Liu G, Yin XM. Homeostatic Role of Autophagy in Hepatocytes. Semin Liver Dis. 2018;38:308-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 184. | Hazari Y, Bravo-San Pedro JM, Hetz C, Galluzzi L, Kroemer G. Autophagy in hepatic adaptation to stress. J Hepatol. 2020;72:183-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 185. | Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 372] [Article Influence: 62.0] [Reference Citation Analysis (0)] |
| 186. | Eslam M, Sanyal AJ, George J; International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020; 158: 1999-2014. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1188] [Cited by in F6Publishing: 1768] [Article Influence: 442.0] [Reference Citation Analysis (1)] |
| 187. | Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, Zelber-Sagi S, Wai-Sun Wong V, Dufour JF, Schattenberg JM, Kawaguchi T, Arrese M, Valenti L, Shiha G, Tiribelli C, Yki-Järvinen H, Fan JG, Grønbæk H, Yilmaz Y, Cortez-Pinto H, Oliveira CP, Bedossa P, Adams LA, Zheng MH, Fouad Y, Chan WK, Mendez-Sanchez N, Ahn SH, Castera L, Bugianesi E, Ratziu V, George J. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J Hepatol. 2020;73:202-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1239] [Cited by in F6Publishing: 2165] [Article Influence: 541.3] [Reference Citation Analysis (0)] |
| 188. | Marra F, Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J Hepatol. 2018;68:280-295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 411] [Cited by in F6Publishing: 526] [Article Influence: 87.7] [Reference Citation Analysis (0)] |
| 189. | Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908-922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1376] [Cited by in F6Publishing: 2401] [Article Influence: 400.2] [Reference Citation Analysis (0)] |
| 190. | Hammoutene A, Rautou PE. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J Hepatol. 2019;70:1278-1291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 194] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 191. | Cingolani F, Czaja MJ. Regulation and Functions of Autophagic Lipolysis. Trends Endocrinol Metab. 2016;27:696-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 192. | Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 379] [Cited by in F6Publishing: 394] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 193. | Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374-3379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1134] [Cited by in F6Publishing: 1137] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 194. | Tomaipitinca L, Mandatori S, Mancinelli R, Giulitti F, Petrungaro S, Moresi V, Facchiano A, Ziparo E, Gaudio E, Giampietri C. The Role of Autophagy in Liver Epithelial Cells and Its Impact on Systemic Homeostasis. Nutrients. 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 195. | Kurahashi T, Hamashima S, Shirato T, Lee J, Homma T, Kang ES, Fujii J. An SOD1 deficiency enhances lipid droplet accumulation in the fasted mouse liver by aborting lipophagy. Biochem Biophys Res Commun. 2015;467:866-871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 196. | Rodriguez-Navarro JA, Kaushik S, Koga H, Dall'Armi C, Shui G, Wenk MR, Di Paolo G, Cuervo AM. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2012;109:E705-E714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 155] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 197. | Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484-31492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 313] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 198. | Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052-3065. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 338] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 199. | Liu K, Lou J, Wen T, Yin J, Xu B, Ding W, Wang A, Liu D, Zhang C, Chen D, Li N. Depending on the stage of hepatosteatosis, p53 causes apoptosis primarily through either DRAM-induced autophagy or BAX. Liver Int. 2013;33:1566-1574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 200. | Kovsan J, Blüher M, Tarnovscki T, Klöting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schön MR, Greenberg AS, Elazar Z, Bashan N, Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268-E277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 250] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 201. | Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 922] [Cited by in F6Publishing: 1010] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 202. | Nuñez CE, Rodrigues VS, Gomes FS, Moura RF, Victorio SC, Bombassaro B, Chaim EA, Pareja JC, Geloneze B, Velloso LA, Araujo EP. Defective regulation of adipose tissue autophagy in obesity. Int J Obes (Lond). 2013;37:1473-1480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 203. | Liu Y, Palanivel R, Rai E, Park M, Gabor TV, Scheid MP, Xu A, Sweeney G. Adiponectin stimulates autophagy and reduces oxidative stress to enhance insulin sensitivity during high-fat diet feeding in mice. Diabetes. 2015;64:36-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 167] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 204. | Lavallard VJ, Meijer AJ, Codogno P, Gual P. Autophagy, signaling and obesity. Pharmacol Res. 2012;66:513-525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 205. | Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87:99-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 605] [Cited by in F6Publishing: 624] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 206. | Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, Nowotny P, Waldhäusl W, Marette A, Roden M. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes. 2005;54:2674-2684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 270] [Cited by in F6Publishing: 280] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 207. | Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900-917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1987] [Cited by in F6Publishing: 2115] [Article Influence: 151.1] [Reference Citation Analysis (0)] |
| 208. | Codogno P, Meijer AJ. Autophagy: a potential link between obesity and insulin resistance. Cell Metab. 2010;11:449-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 209. | Inami Y, Yamashina S, Izumi K, Ueno T, Tanida I, Ikejima K, Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun. 2011;412:618-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 210. | Fukuo Y, Yamashina S, Sonoue H, Arakawa A, Nakadera E, Aoyama T, Uchiyama A, Kon K, Ikejima K, Watanabe S. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol Res. 2014;44:1026-1036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 211. | Wang X, Zhang X, Chu ESH, Chen X, Kang W, Wu F, To KF, Wong VWS, Chan HLY, Chan MTV, Sung JJY, Wu WKK, Yu J. Defective lysosomal clearance of autophagosomes and its clinical implications in nonalcoholic steatohepatitis. FASEB J. 2018;32:37-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 212. | Mizunoe Y, Sudo Y, Okita N, Hiraoka H, Mikami K, Narahara T, Negishi A, Yoshida M, Higashibata R, Watanabe S, Kaneko H, Natori D, Furuichi T, Yasukawa H, Kobayashi M, Higami Y. Involvement of lysosomal dysfunction in autophagosome accumulation and early pathologies in adipose tissue of obese mice. Autophagy. 2017;13:642-653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 213. | Ju L, Han J, Zhang X, Deng Y, Yan H, Wang C, Li X, Chen S, Alimujiang M, Li X, Fang Q, Yang Y, Jia W. Obesity-associated inflammation triggers an autophagy-lysosomal response in adipocytes and causes degradation of perilipin 1. Cell Death Dis. 2019;10:121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 214. | Mizunoe Y, Kobayashi M, Tagawa R, Nakagawa Y, Shimano H, Higami Y. Association between Lysosomal Dysfunction and Obesity-Related Pathology: A Key Knowledge to Prevent Metabolic Syndrome. Int J Mol Sci. 2019;20:3688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 215. | Qian Q, Zhang Z, Li M, Savage K, Cheng D, Rauckhorst AJ, Ankrum JA, Taylor EB, Ding WX, Xiao Y, Cao HJ, Yang L. Hepatic Lysosomal iNOS Activity Impairs Autophagy in Obesity. Cell Mol Gastroenterol Hepatol. 2019;8:95-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 216. | Wu H, Wang Y, Li W, Chen H, Du L, Liu D, Wang X, Xu T, Liu L, Chen Q. Deficiency of mitophagy receptor FUNDC1 impairs mitochondrial quality and aggravates dietary-induced obesity and metabolic syndrome. Autophagy. 2019;15:1882-1898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 217. | Su Z, Nie Y, Huang X, Zhu Y, Feng B, Tang L, Zheng G. Mitophagy in Hepatic Insulin Resistance: Therapeutic Potential and Concerns. Front Pharmacol. 2019;10:1193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 218. | Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, Lee MS. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19:83-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 550] [Cited by in F6Publishing: 615] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 219. | Zhu S, Wu Y, Ye X, Ma L, Qi J, Yu D, Wei Y, Lin G, Ren G, Li D. FGF21 ameliorates nonalcoholic fatty liver disease by inducing autophagy. Mol Cell Biochem. 2016;420:107-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 220. | Shen J, Chan HL, Wong GL, Choi PC, Chan AW, Chan HY, Chim AM, Yeung DK, Chan FK, Woo J, Yu J, Chu WC, Wong VW. Non-invasive diagnosis of non-alcoholic steatohepatitis by combined serum biomarkers. J Hepatol. 2012;56:1363-1370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 221. | Adams AC, Halstead CA, Hansen BC, Irizarry AR, Martin JA, Myers SR, Reynolds VL, Smith HW, Wroblewski VJ, Kharitonenkov A. LY2405319, an Engineered FGF21 Variant, Improves the Metabolic Status of Diabetic Monkeys. PLoS One. 2013;8:e65763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 222. | Kharitonenkov A, Beals JM, Micanovic R, Strifler BA, Rathnachalam R, Wroblewski VJ, Li S, Koester A, Ford AM, Coskun T, Dunbar JD, Cheng CC, Frye CC, Bumol TF, Moller DE. Rational design of a fibroblast growth factor 21-based clinical candidate, LY2405319. PLoS One. 2013;8:e58575. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 223. | Gaich G, Chien JY, Fu H, Glass LC, Deeg MA, Holland WL, Kharitonenkov A, Bumol T, Schilske HK, Moller DE. The effects of LY2405319, an FGF21 analog, in obese human subjects with type 2 diabetes. Cell Metab. 2013;18:333-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 632] [Cited by in F6Publishing: 676] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 224. | Romero M, Zorzano A. Role of autophagy in the regulation of adipose tissue biology. Cell Cycle. 2019;18:1435-1445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 225. | Wang L, Liu X, Nie J, Zhang J, Kimball SR, Zhang H, Zhang WJ, Jefferson LS, Cheng Z, Ji Q, Shi Y. ALCAT1 controls mitochondrial etiology of fatty liver diseases, linking defective mitophagy to steatosis. Hepatology. 2015;61:486-496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 226. | Sinha RA, Yen PM. Thyroid hormone-mediated autophagy and mitochondrial turnover in NAFLD. Cell Biosci. 2016;6:46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 227. | Chi HC, Tsai CY, Tsai MM, Yeh CT, Lin KH. Molecular functions and clinical impact of thyroid hormone-triggered autophagy in liver-related diseases. J Biomed Sci. 2019;26:24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 228. | Pang L, Liu K, Liu D, Lv F, Zang Y, Xie F, Yin J, Shi Y, Wang Y, Chen D. Differential effects of reticulophagy and mitophagy on nonalcoholic fatty liver disease. Cell Death Dis. 2018;9:90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 229. | Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, Yanagawa T, Okamoto K, Iijima M, Sesaki H. Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in Parkin-Independent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease. Cell Metab 2018; 28: 588-604. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 230. | Lee K, Haddad A, Osme A, Kim C, Borzou A, Ilchenko S, Allende D, Dasarathy S, McCullough A, Sadygov RG, Kasumov T. Hepatic Mitochondrial Defects in a Nonalcoholic Fatty Liver Disease Mouse Model Are Associated with Increased Degradation of Oxidative Phosphorylation Subunits. Mol Cell Proteomics. 2018;17:2371-2386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 231. | Shao N, Yu XY, Ma XF, Lin WJ, Hao M, Kuang HY. Exenatide Delays the Progression of Nonalcoholic Fatty Liver Disease in C57BL/6 Mice, Which May Involve Inhibition of the NLRP3 Inflammasome through the Mitophagy Pathway. Gastroenterol Res Pract. 2018;2018:1864307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 232. | Li R, Xin T, Li D, Wang C, Zhu H, Zhou H. Therapeutic effect of Sirtuin 3 on ameliorating nonalcoholic fatty liver disease: The role of the ERK-CREB pathway and Bnip3-mediated mitophagy. Redox Biol. 2018;18:229-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 247] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 233. | Yu X, Hao M, Liu Y, Ma X, Lin W, Xu Q, Zhou H, Shao N, Kuang H. Liraglutide ameliorates non-alcoholic steatohepatitis by inhibiting NLRP3 inflammasome and pyroptosis activation via mitophagy. Eur J Pharmacol. 2019;864:172715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 234. | Zhou T, Chang L, Luo Y, Zhou Y, Zhang J. Mst1 inhibition attenuates non-alcoholic fatty liver disease via reversing Parkin-related mitophagy. Redox Biol. 2019;21:101120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 235. | Niso-Santano M, Malik SA, Pietrocola F, Bravo-San Pedro JM, Mariño G, Cianfanelli V, Ben-Younès A, Troncoso R, Markaki M, Sica V, Izzo V, Chaba K, Bauvy C, Dupont N, Kepp O, Rockenfeller P, Wolinski H, Madeo F, Lavandero S, Codogno P, Harper F, Pierron G, Tavernarakis N, Cecconi F, Maiuri MC, Galluzzi L, Kroemer G. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025-1041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 236. | Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329-3339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 237. | Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding WX. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 230] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 238. | Deng X, Pan X, Cheng C, Liu B, Zhang H, Zhang Y, Xu K. Regulation of SREBP-2 intracellular trafficking improves impaired autophagic flux and alleviates endoplasmic reticulum stress in NAFLD. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:337-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 239. | Maus M, Cuk M, Patel B, Lian J, Ouimet M, Kaufmann U, Yang J, Horvath R, Hornig-Do HT, Chrzanowska-Lightowlers ZM, Moore KJ, Cuervo AM, Feske S. Store-Operated Ca2+ Entry Controls Induction of Lipolysis and the Transcriptional Reprogramming to Lipid Metabolism. Cell Metab. 2017;25:698-712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 240. | Li S, Dou X, Ning H, Song Q, Wei W, Zhang X, Shen C, Li J, Sun C, Song Z. Sirtuin 3 acts as a negative regulator of autophagy dictating hepatocyte susceptibility to lipotoxicity. Hepatology. 2017;66:936-952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 241. | Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y, Ilkayeva OR, Gooding J, Ching J, Zhou J, Martinez L, Xie S, Bay BH, Summers SA, Newgard CB, Yen PM. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology. 2014;59:1366-1380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 248] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 242. | Ding WX. Drinking coffee burns hepatic fat by inducing lipophagy coupled with mitochondrial β-oxidation. Hepatology. 2014;59:1235-1238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 243. | Molloy JW, Calcagno CJ, Williams CD, Jones FJ, Torres DM, Harrison SA. Association of coffee and caffeine consumption with fatty liver disease, nonalcoholic steatohepatitis, and degree of hepatic fibrosis. Hepatology. 2012;55:429-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 212] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 244. | Anty R, Marjoux S, Iannelli A, Patouraux S, Schneck AS, Bonnafous S, Gire C, Amzolini A, Ben-Amor I, Saint-Paul MC, Mariné-Barjoan E, Pariente A, Gugenheim J, Gual P, Tran A. Regular coffee but not espresso drinking is protective against fibrosis in a cohort mainly composed of morbidly obese European women with NAFLD undergoing bariatric surgery. J Hepatol. 2012;57:1090-1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 245. | Chen R, Wang Q, Song S, Liu F, He B, Gao X. Protective role of autophagy in methionine-choline deficient diet-induced advanced nonalcoholic steatohepatitis in mice. Eur J Pharmacol. 2016;770:126-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 246. | Ji G, Wang Y, Deng Y, Li X, Jiang Z. Resveratrol ameliorates hepatic steatosis and inflammation in methionine/choline-deficient diet-induced steatohepatitis through regulating autophagy. Lipids Health Dis. 2015;14:134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 247. | Ding S, Jiang J, Zhang G, Bu Y, Zhang G, Zhao X. Resveratrol and caloric restriction prevent hepatic steatosis by regulating SIRT1-autophagy pathway and alleviating endoplasmic reticulum stress in high-fat diet-fed rats. PLoS One. 2017;12:e0183541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 248. | Xiao L, Liang S, Ge L, Qiu S, Wan H, Wu S, Fei J, Peng S, Zeng X. Si-Wei-Qing-Gan-Tang Improves Non-Alcoholic Steatohepatitis by Modulating the Nuclear Factor-κB Signal Pathway and Autophagy in Methionine and Choline Deficient Diet-Fed Rats. Front Pharmacol. 2020;11:530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 249. | Wang X, Zhang ZF, Zheng GH, Wang AM, Sun CH, Qin SP, Zhuang J, Lu J, Ma DF, Zheng YL. The Inhibitory Effects of Purple Sweet Potato Color on Hepatic Inflammation Is Associated with Restoration of NAD⁺ Levels and Attenuation of NLRP3 Inflammasome Activation in High-Fat-Diet-Treated Mice. Molecules. 2017;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 250. | Kim HM, Kim Y, Lee ES, Huh JH, Chung CH. Caffeic acid ameliorates hepatic steatosis and reduces ER stress in high fat diet-induced obese mice by regulating autophagy. Nutrition. 2018;55-56:63-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 251. | Elmansi AM, El-Karef AA, Shishtawy MMEl, Eissa LA. Hepatoprotective Effect of Curcumin on Hepatocellular Carcinoma Through Autophagic and Apoptic Pathways. Ann Hepatol. 2017;16:607-618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 252. | Liu C, Liu L, Zhu HD, Sheng JQ, Wu XL, He XX, Tian DA, Liao JZ, Li PY. Celecoxib alleviates nonalcoholic fatty liver disease by restoring autophagic flux. Sci Rep. 2018;8:4108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 253. | Alexaki A, Gupta SD, Majumder S, Kono M, Tuymetova G, Harmon JM, Dunn TM, Proia RL. Autophagy regulates sphingolipid levels in the liver. J Lipid Res. 2014;55:2521-2531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 254. | Menikdiwela KR, Ramalingam L, Rasha F, Wang S, Dufour JM, Kalupahana NS, Sunahara KKS, Martins JO, Moustaid-Moussa N. Autophagy in metabolic syndrome: breaking the wheel by targeting the renin-angiotensin system. Cell Death Dis. 2020;11:87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 255. | Tanaka S, Hikita H, Tatsumi T, Sakamori R, Nozaki Y, Sakane S, Shiode Y, Nakabori T, Saito Y, Hiramatsu N, Tabata K, Kawabata T, Hamasaki M, Eguchi H, Nagano H, Yoshimori T, Takehara T. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology. 2016;64:1994-2014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 245] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 256. | Yan H, Gao Y, Zhang Y. Inhibition of JNK suppresses autophagy and attenuates insulin resistance in a rat model of nonalcoholic fatty liver disease. Mol Med Rep. 2017;15:180-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 257. | Lavallard VJ, Gual P. Autophagy and non-alcoholic fatty liver disease. Biomed Res Int. 2014;2014:120179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 258. | Mao Y, Yu F, Wang J, Guo C, Fan X. Autophagy: a new target for nonalcoholic fatty liver disease therapy. Hepat Med. 2016;8:27-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 259. | Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 228] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 260. | Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759-770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 493] [Cited by in F6Publishing: 469] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 261. | Schneider JL, Villarroya J, Diaz-Carretero A, Patel B, Urbanska AM, Thi MM, Villarroya F, Santambrogio L, Cuervo AM. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell. 2015;14:249-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 262. | Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467-2474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2702] [Cited by in F6Publishing: 2771] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 263. | Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ; Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313-1321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6807] [Cited by in F6Publishing: 7652] [Article Influence: 402.7] [Reference Citation Analysis (5)] |
| 264. | Bedossa P, Poitou C, Veyrie N, Bouillot JL, Basdevant A, Paradis V, Tordjman J, Clement K. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56:1751-1759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 526] [Cited by in F6Publishing: 594] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 265. | Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47:2026-2035. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 266. | González-Rodríguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillón J, Lo Iacono O, Corazzari M, Fimia GM, Piacentini M, Muntané J, Boscá L, García-Monzón C, Martín-Sanz P, Valverde ÁM. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 433] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 267. | Li P, He K, Li J, Liu Z, Gong J. The role of Kupffer cells in hepatic diseases. Mol Immunol. 2017;85:222-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 138] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 268. | Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol. 2017;66:1300-1312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 504] [Cited by in F6Publishing: 629] [Article Influence: 89.9] [Reference Citation Analysis (0)] |
| 269. | Thomas H. NAFLD: A critical role for the NLRP3 inflammasome in NASH. Nat Rev Gastroenterol Hepatol. 2017;14:197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 270. | Feldstein AE, Werneburg NW, Canbay A, Guicciardi ME, Bronk SF, Rydzewski R, Burgart LJ, Gores GJ. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology. 2004;40:185-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 586] [Cited by in F6Publishing: 589] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 271. | Tang Y, Cao G, Min X, Wang T, Sun S, Du X, Zhang W. Cathepsin B inhibition ameliorates the non-alcoholic steatohepatitis through suppressing caspase-1 activation. J Physiol Biochem. 2018;74:503-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 272. | Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW Jr. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816-830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 346] [Cited by in F6Publishing: 367] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 273. | Grijalva A, Xu X, Ferrante AW Jr. Autophagy Is Dispensable for Macrophage-Mediated Lipid Homeostasis in Adipose Tissue. Diabetes. 2016;65:967-980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 274. | Bieghs V, Hendrikx T, van Gorp PJ, Verheyen F, Guichot YD, Walenbergh SM, Jeurissen ML, Gijbels M, Rensen SS, Bast A, Plat J, Kalhan SC, Koek GH, Leitersdorf E, Hofker MH, Lütjohann D, Shiri-Sverdlov R. The cholesterol derivative 27-hydroxycholesterol reduces steatohepatitis in mice. Gastroenterology 2013; 144: 167-178. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 275. | Houben T, Oligschlaeger Y, Hendrikx T, Bitorina AV, Walenbergh SMA, van Gorp PJ, Gijbels MJJ, Friedrichs S, Plat J, Schaap FG, Lütjohann D, Hofker MH, Shiri-Sverdlov R. Cathepsin D regulates lipid metabolism in murine steatohepatitis. Sci Rep. 2017;7:3494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 276. | Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287:356-359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4680] [Cited by in F6Publishing: 4448] [Article Influence: 202.2] [Reference Citation Analysis (0)] |
| 277. | Noureddin M, Yates KP, Vaughn IA, Neuschwander-Tetri BA, Sanyal AJ, McCullough A, Merriman R, Hameed B, Doo E, Kleiner DE, Behling C, Loomba R; NASH CRN. Clinical and histological determinants of nonalcoholic steatohepatitis and advanced fibrosis in elderly patients. Hepatology. 2013;58:1644-1654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 278. | Li Z, Berk M, McIntyre TM, Gores GJ, Feldstein AE. The lysosomal-mitochondrial axis in free fatty acid-induced hepatic lipotoxicity. Hepatology. 2008;47:1495-1503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 279. | Sunny NE, Bril F, Cusi K. Mitochondrial Adaptation in Nonalcoholic Fatty Liver Disease: Novel Mechanisms and Treatment Strategies. Trends Endocrinol Metab. 2017;28:250-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 218] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 280. | Simões ICM, Fontes A, Pinton P, Zischka H, Wieckowski MR. Mitochondria in non-alcoholic fatty liver disease. Int J Biochem Cell Biol. 2018;95:93-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 163] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 281. | Hammoutene A, Lasselin J, Vion A, Colnot N, Paradis V, Lotersztajn S, Boulanger C, Rautou P. Defective autophagy in liver sinusoidal endothelial cells promotes non alcoholic steatohepatitis and fibrosis development. J Hepatol. 2018;68:S29. [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 282. | Hammoutene A, Biquard L, Lasselin J, Kheloufi M, Tanguy M, Vion AC, Mérian J, Colnot N, Loyer X, Tedgui A, Codogno P, Lotersztajn S, Paradis V, Boulanger CM, Rautou PE. A defect in endothelial autophagy occurs in patients with non-alcoholic steatohepatitis and promotes inflammation and fibrosis. J Hepatol. 2020;72:528-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 101] [Article Influence: 25.3] [Reference Citation Analysis (1)] |
| 283. | Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 362] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 284. | Zhao GN, Zhang P, Gong J, Zhang XJ, Wang PX, Yin M, Jiang Z, Shen LJ, Ji YX, Tong J, Wang Y, Wei QF, Wang Y, Zhu XY, Zhang X, Fang J, Xie Q, She ZG, Wang Z, Huang Z, Li H. Tmbim1 is a multivesicular body regulator that protects against non-alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat Med. 2017;23:742-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 285. | Zhang X, Wu WK, Xu W, Man K, Wang X, Han J, Leung WY, Wu R, Liu K, Yu J. C-X-C Motif Chemokine 10 Impairs Autophagy and Autolysosome Formation in Non-alcoholic Steatohepatitis. Theranostics. 2017;7:2822-2836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 286. | Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol Res Health. 2006;29:245-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 287. | Dey A, Cederbaum AI. Alcohol and oxidative liver injury. Hepatology. 2006;43:S63-S74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 398] [Cited by in F6Publishing: 412] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 288. | Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740-1752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 376] [Cited by in F6Publishing: 411] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 289. | Kong X, Yang Y, Ren L, Shao T, Li F, Zhao C, Liu L, Zhang H, McClain CJ, Feng W. Activation of autophagy attenuates EtOH-LPS-induced hepatic steatosis and injury through MD2 associated TLR4 signaling. Sci Rep. 2017;7:9292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 290. | Thomes PG, Ehlers RA, Trambly CS, Clemens DL, Fox HS, Tuma DJ, Donohue TM. Multilevel regulation of autophagosome content by ethanol oxidation in HepG2 cells. Autophagy. 2013;9:63-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 291. | Wu D, Wang X, Zhou R, Yang L, Cederbaum AI. Alcohol steatosis and cytotoxicity: the role of cytochrome P4502E1 and autophagy. Free Radic Biol Med. 2012;53:1346-1357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 292. | Sid B, Verrax J, Calderon PB. Role of AMPK activation in oxidative cell damage: Implications for alcohol-induced liver disease. Biochem Pharmacol. 2013;86:200-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 293. | Ding WX, Li M, Yin XM. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7:248-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 294. | Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749-1760. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1509] [Cited by in F6Publishing: 1576] [Article Influence: 92.7] [Reference Citation Analysis (0)] |
| 295. | Thomes PG, Trambly CS, Thiele GM, Duryee MJ, Fox HS, Haorah J, Donohue TM Jr. Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochem Biophys Res Commun. 2012;417:262-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 296. | Yeon JE, Califano S, Xu J, Wands JR, De La Monte SM. Potential role of PTEN phosphatase in ethanol-impaired survival signaling in the liver. Hepatology. 2003;38:703-714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 297. | Shulga N, Hoek JB, Pastorino JG. Elevated PTEN levels account for the increased sensitivity of ethanol-exposed cells to tumor necrosis factor-induced cytotoxicity. J Biol Chem. 2005;280:9416-9424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 298. | Liuzzi JP, Yoo C. Role of zinc in the regulation of autophagy during ethanol exposure in human hepatoma cells. Biol Trace Elem Res. 2013;156:350-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 299. | Wu D, Wang X, Zhou R, Cederbaum A. CYP2E1 enhances ethanol-induced lipid accumulation but impairs autophagy in HepG2 E47 cells. Biochem Biophys Res Commun. 2010;402:116-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 300. | Guo R, Xu X, Babcock SA, Zhang Y, Ren J. Aldehyde dedydrogenase-2 plays a beneficial role in ameliorating chronic alcohol-induced hepatic steatosis and inflammation through regulation of autophagy. J Hepatol. 2015;62:647-656. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 301. | Denaës T, Lodder J, Chobert MN, Ruiz I, Pawlotsky JM, Lotersztajn S, Teixeira-Clerc F. The Cannabinoid Receptor 2 Protects Against Alcoholic Liver Disease Via a Macrophage Autophagy-Dependent Pathway. Sci Rep. 2016;6:28806. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 302. | Thomes PG, Trambly CS, Fox HS, Tuma DJ, Donohue TM Jr. Acute and Chronic Ethanol Administration Differentially Modulate Hepatic Autophagy and Transcription Factor EB. Alcohol Clin Exp Res. 2015;39:2354-2363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 303. | Ni HM, Du K, You M, Ding WX. Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am J Pathol. 2013;183:1815-1825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 304. | Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479-2487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 786] [Cited by in F6Publishing: 857] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 305. | Eid N, Ito Y, Maemura K, Otsuki Y. Elevated autophagic sequestration of mitochondria and lipid droplets in steatotic hepatocytes of chronic ethanol-treated rats: an immunohistochemical and electron microscopic study. J Mol Histol. 2013;44:311-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 306. | Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X, Dong XC, Yin XM. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993-999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 325] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 307. | Kharbanda KK, McVicker DL, Zetterman RK, Donohue TM Jr. Ethanol consumption reduces the proteolytic capacity and protease activities of hepatic lysosomes. Biochim Biophys Acta. 1995;1245:421-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 60] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 308. | Chao X, Wang S, Zhao K, Li Y, Williams JA, Li T, Chavan H, Krishnamurthy P, He XC, Li L, Ballabio A, Ni HM, Ding WX. Impaired TFEB-Mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-Induced Liver Injury and Steatosis in Mice. Gastroenterology 2018; 155: 865-879. e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 218] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 309. | Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123-1134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 316] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 310. | You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798-1808. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 345] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 311. | Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1546] [Cited by in F6Publishing: 1792] [Article Influence: 128.0] [Reference Citation Analysis (0)] |
| 312. | Lu Y, Cederbaum AI. Autophagy Protects against CYP2E1/Chronic Ethanol-Induced Hepatotoxicity. Biomolecules. 2015;5:2659-2674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 313. | Liu L, Xie P, Li W, Wu Y, An W. Augmenter of Liver Regeneration Protects against Ethanol-Induced Acute Liver Injury by Promoting Autophagy. Am J Pathol. 2019;189:552-567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 314. | Yan S, Zhou J, Chen X, Dong Z, Yin XM. Diverse Consequences in Liver Injury in Mice with Different Autophagy Functional Status Treated with Alcohol. Am J Pathol. 2019;189:1744-1762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 315. | Williams JA, Ding WX. A Mechanistic Review of Mitophagy and Its Role in Protection against Alcoholic Liver Disease. Biomolecules. 2015;5:2619-2642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 316. | Eid N, Ito Y, Otsuki Y. Triggering of Parkin Mitochondrial Translocation in Mitophagy: Implications for Liver Diseases. Front Pharmacol. 2016;7:100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 317. | Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107-39114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 318. | Harada M. Autophagy is involved in the elimination of intracellular inclusions, Mallory-Denk bodies, in hepatocytes. Med Mol Morphol. 2010;43:13-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 319. | Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782:764-774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 320. | Schulze RJ, Rasineni K, Weller SG, Schott MB, Schroeder B, Casey CA, McNiven MA. Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol Commun. 2017;1:140-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 321. | Rasineni K, Donohue TM Jr, Thomes PG, Yang L, Tuma DJ, McNiven MA, Casey CA. Ethanol-induced steatosis involves impairment of lipophagy, associated with reduced Dynamin2 activity. Hepatol Commun. 2017;1:501-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 322. | Eid N, Ito Y, Horibe A, Otsuki Y. Ethanol-induced mitophagy in liver is associated with activation of the PINK1-Parkin pathway triggered by oxidative DNA damage. Histol Histopathol. 2016;31:1143-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 19] [Reference Citation Analysis (0)] |
| 323. | Eid N, Ito Y, Horibe A, Otsuki Y, Kondo Y. Ethanol-Induced Mitochondrial Damage in Sertoli Cells is Associated with Parkin Overexpression and Activation of Mitophagy. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 324. | Williams JA, Ni HM, Ding Y, Ding WX. Parkin regulates mitophagy and mitochondrial function to protect against alcohol-induced liver injury and steatosis in mice. Am J Physiol Gastrointest Liver Physiol. 2015;309:G324-G340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 133] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 325. | Eid N, Ito Y, Otsuki Y. Mitophagy in steatotic hepatocytes of ethanol-treated wild-type and Parkin knockout mice. Am J Physiol Gastrointest Liver Physiol. 2015;309:G513-G514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 326. | Yu X, Xu Y, Zhang S, Sun J, Liu P, Xiao L, Tang Y, Liu L, Yao P. Quercetin Attenuates Chronic Ethanol-Induced Hepatic Mitochondrial Damage through Enhanced Mitophagy. Nutrients. 2016;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 327. | Zhou H, Wang S, Hu S, Chen Y, Ren J. ER-Mitochondria Microdomains in Cardiac Ischemia-Reperfusion Injury: A Fresh Perspective. Front Physiol. 2018;9:755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 328. | Zhou H, Zhu P, Wang J, Toan S, Ren J. DNA-PKcs promotes alcohol-related liver disease by activating Drp1-related mitochondrial fission and repressing FUNDC1-required mitophagy. Signal Transduct Target Ther. 2019;4:56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 329. | Ilyas G, Cingolani F, Zhao E, Tanaka K, Czaja MJ. Decreased Macrophage Autophagy Promotes Liver Injury and Inflammation from Alcohol. Alcohol Clin Exp Res. 2019;43:1403-1413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 330. | Liang S, Zhong Z, Kim SY, Uchiyama R, Roh YS, Matsushita H, Gottlieb RA, Seki E. Murine macrophage autophagy protects against alcohol-induced liver injury by degrading interferon regulatory factor 1 (IRF1) and removing damaged mitochondria. J Biol Chem. 2019;294:12359-12369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 331. | Xie ZY, Xiao ZH, Wang FF. Inhibition of autophagy reverses alcohol-induced hepatic stellate cells activation through activation of Nrf2-Keap1-ARE signaling pathway. Biochimie. 2018;147:55-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 332. | Szabo G, Kamath PS, Shah VH, Thursz M, Mathurin P; EASL-AASLD Joint Meeting. Alcohol-Related Liver Disease: Areas of Consensus, Unmet Needs and Opportunities for Further Study. Hepatology. 2019;69:2271-2283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 333. | You Y, Li WZ, Zhang S, Hu B, Li YX, Li HD, Tang HH, Li QW, Guan YY, Liu LX, Bao WL, Shen X. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy. J Hepatol. 2018;69:129-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 334. | Cai Y, Jogasuria A, Yin H, Xu MJ, Hu X, Wang J, Kim C, Wu J, Lee K, Gao B, You M. The Detrimental Role Played by Lipocalin-2 in Alcoholic Fatty Liver in Mice. Am J Pathol. 2016;186:2417-2428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 335. | Han W, Fu X, Xie J, Meng Z, Gu Y, Wang X, Li L, Pan H, Huang W. MiR-26a enhances autophagy to protect against ethanol-induced acute liver injury. J Mol Med (Berl). 2015;93:1045-1055. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 336. | Babuta M, Furi I, Bala S, Bukong TN, Lowe P, Catalano D, Calenda C, Kodys K, Szabo G. Dysregulated Autophagy and Lysosome Function Are Linked to Exosome Production by Micro-RNA 155 in Alcoholic Liver Disease. Hepatology. 2019;70:2123-2141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 337. | Thomes PG, Rasineni K, Yang L, Donohue TM Jr, Kubik JL, McNiven MA, Casey CA. Ethanol withdrawal mitigates fatty liver by normalizing lipid catabolism. Am J Physiol Gastrointest Liver Physiol. 2019;316:G509-G518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 338. | Kirchgesner T, Danse E. Drink responsibly! JBR-BTR. 2014;97:44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 339. | Thiele M, Rausch V, Fluhr G, Kjærgaard M, Piecha F, Mueller J, Straub BK, Lupșor-Platon M, De-Ledinghen V, Seitz HK, Detlefsen S, Madsen B, Krag A, Mueller S. Controlled attenuation parameter and alcoholic hepatic steatosis: Diagnostic accuracy and role of alcohol detoxification. J Hepatol. 2018;68:1025-1032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 340. | Yan S, Khambu B, Hong H, Liu G, Huda N, Yin XM. Autophagy, Metabolism, and Alcohol-Related Liver Disease: Novel Modulators and Functions. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 341. | Yang L, Yang C, Thomes PG, Kharbanda KK, Casey CA, McNiven MA, Donohue TM Jr. Lipophagy and Alcohol-Induced Fatty Liver. Front Pharmacol. 2019;10:495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 342. | Chao X, Ding WX. Role and mechanisms of autophagy in alcohol-induced liver injury. Adv Pharmacol. 2019;85:109-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 343. | Tang L, Yang F, Fang Z, Hu C. Resveratrol Ameliorates Alcoholic Fatty Liver by Inducing Autophagy. Am J Chin Med. 2016;44:1207-1220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 344. | Atef MM, Hafez YM, Alshenawy HA, Emam MN. Ameliorative effects of autophagy inducer, simvastatin on alcohol-induced liver disease in a rat model. J Cell Biochem. 2018;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 345. | Guo X, Cui R, Zhao J, Mo R, Peng L, Yan M. Corosolic acid protects hepatocytes against ethanol-induced damage by modulating mitogen-activated protein kinases and activating autophagy. Eur J Pharmacol. 2016;791:578-588. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 346. | Li Y, Chen M, Wang J, Guo X, Xiao L, Liu P, Liu L, Tang Y, Yao P. Quercetin ameliorates autophagy in alcohol liver disease associated with lysosome through mTOR-TFEB pathway. J Funct Foods. 2019;52:177-185. [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 347. | Shi X, Sun R, Zhao Y, Fu R, Wang R, Zhao H, Wang Z, Tang F, Zhang N, Tian X, Yao J. Promotion of autophagosome–lysosome fusion via salvianolic acid A-mediated SIRT1 upregulation ameliorates alcoholic liver disease. RSC Adv. 2018;8:20411-20422. [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 348. | Lemasters JJ, Zhong Z. Mitophagy in hepatocytes: Types, initiators and role in adaptive ethanol metabolism. Liver Res. 2018;2:125-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 349. | Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011;415:68-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 350. | Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, Liu W. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 131] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 351. | Cho HK, Cheong KJ, Kim HY, Cheong J. Endoplasmic reticulum stress induced by hepatitis B virus X protein enhances cyclo-oxygenase 2 expression via activating transcription factor 4. Biochem J. 2011;435:431-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 352. | Li B, Gao B, Ye L, Han X, Wang W, Kong L, Fang X, Zeng Y, Zheng H, Li S, Wu Z, Ye L. Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 2007;124:44-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 113] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 353. | Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387-7392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 128] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 354. | Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023-2032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 251] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 355. | Na B, Huang Z, Wang Q, Qi Z, Tian Y, Lu CC, Yu J, Hanes MA, Kakar S, Huang EJ, Ou JH, Liu L, Yen TS. Transgenic expression of entire hepatitis B virus in mice induces hepatocarcinogenesis independent of chronic liver injury. PLoS One. 2011;6:e26240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 356. | Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319-6333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 357. | Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 192] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 358. | Zhong L, Shu W, Dai W, Gao B, Xiong S. Reactive Oxygen Species-Mediated c-Jun NH2-Terminal Kinase Activation Contributes to Hepatitis B Virus X Protein-Induced Autophagy via Regulation of the Beclin-1/Bcl-2 Interaction. J Virol. 2017;91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 359. | Fu S, Wang J, Hu X, Zhou RR, Fu Y, Tang D, Kang R, Huang Y, Sun L, Li N, Fan XG. Crosstalk between hepatitis B virus X and high-mobility group box 1 facilitates autophagy in hepatocytes. Mol Oncol. 2018;12:322-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 360. | Cheng LS, Li J, Liu Y, Wang FP, Wang SQ, She WM, Wu SD, Qi XL, Zhou YP, Jiang W. HMGB1-induced autophagy: a new pathway to maintain Treg function during chronic hepatitis B virus infection. Clin Sci (Lond). 2017;131:381-394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 361. | Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9:e1003722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 223] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 362. | Huang XY, Li D, Chen ZX, Huang YH, Gao WY, Zheng BY, Wang XZ. Hepatitis B Virus X protein elevates Parkin-mediated mitophagy through Lon Peptidase in starvation. Exp Cell Res. 2018;368:75-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 363. | Wang J, Shi Y, Yang H. [Infection with hepatitis B virus enhances basal autophagy]. Wei Sheng Wu Xue Bao. 2010;50:1651-1656. [PubMed] [Cited in This Article: ] |
| 364. | Lin CL, Kao JH. Hepatitis B viral factors and clinical outcomes of chronic hepatitis B. J Biomed Sci. 2008;15:137-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 365. | Xie M, Yang Z, Liu Y, Zheng M. The role of HBV-induced autophagy in HBV replication and HBV related-HCC. Life Sci. 2018;205:107-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 366. | Choi Y, Bowman JW, Jung JU. Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol. 2018;16:341-354. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 342] [Cited by in F6Publishing: 488] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 367. | Sir D, Ou JH. Autophagy in viral replication and pathogenesis. Mol Cells. 2010;29:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 368. | Sir D, Ann DK, Ou JH. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy. 2010;6:548-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 369. | Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383-4388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 240] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 370. | Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453-13456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 371. | Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505-517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 159] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 372. | Luo MX, Wong SH, Chan MT, Yu L, Yu SS, Wu F, Xiao Z, Wang X, Zhang L, Cheng AS, Ng SS, Chan FK, Cho CH, Yu J, Sung JJ, Wu WK. Autophagy Mediates HBx-Induced Nuclear Factor-κB Activation and Release of IL-6, IL-8, and CXCL2 in Hepatocytes. J Cell Physiol. 2015;230:2382-2389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 373. | Hu S, Liu X, Gao Y, Zhou R, Wei M, Dong J, Yan H, Zhao Y. Hepatitis B Virus Inhibits Neutrophil Extracellular Trap Release by Modulating Reactive Oxygen Species Production and Autophagy. J Immunol. 2019;202:805-815. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 374. | Lin Y, Wu C, Wang X, Liu S, Kemper T, Li F, Squire A, Zhu Y, Zhang J, Chen X, Lu M. Synaptosomal-associated protein 29 is required for the autophagic degradation of hepatitis B virus. FASEB J. 2019;33:6023-6034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 375. | Lin Y, Wu C, Wang X, Liu S, Zhao K, Kemper T, Yu H, Li M, Zhang J, Chen M, Zhu Y, Chen X, Lu M. Glucosamine promotes hepatitis B virus replication through its dual effects in suppressing autophagic degradation and inhibiting MTORC1 signaling. Autophagy. 2020;16:548-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 376. | Gracia-Sancho J, Guixé-Muntet S, Hide D, Bosch J. Modulation of autophagy for the treatment of liver diseases. Expert Opin Investig Drugs. 2014;23:965-977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 377. | Döring T, Zeyen L, Bartusch C, Prange R. Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does Not Require Atg8/LC3 Lipidation for Viral Maturation. J Virol. 2018;92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 378. | Shin GC, Kang HS, Lee AR, Kim KH. Hepatitis B virus-triggered autophagy targets TNFRSF10B/death receptor 5 for degradation to limit TNFSF10/TRAIL response. Autophagy. 2016;12:2451-2466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 379. | Khabir M, Aliche AZ, Sureau C, Blanchet M, Labonté P. Hepatitis Delta Virus Alters the Autophagy Process To Promote Its Genome Replication. J Virol. 2020;94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 380. | Khan M, Imam H, Siddiqui A. Subversion of cellular autophagy during virus infection: Insights from hepatitis B and hepatitis C viruses. Liver Res. 2018;2:146-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 381. | Wang L, Ou JH. Hepatitis C virus and autophagy. Biol Chem. 2015;396:1215-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 382. | Chan ST, Ou JJ. Hepatitis C Virus-Induced Autophagy and Host Innate Immune Response. Viruses. 2017;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 383. | Wang J, Kang R, Huang H, Xi X, Wang B, Wang J, Zhao Z. Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression. Autophagy. 2014;10:766-784. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 384. | Huang H, Kang R, Wang J, Luo G, Yang W, Zhao Z. Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy. 2013;9:175-195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 385. | Kim JY, Wang L, Lee J, Ou JJ. Hepatitis C Virus Induces the Localization of Lipid Rafts to Autophagosomes for Its RNA Replication. J Virol. 2017;91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 386. | Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem. 2012;287:18036-18043. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 131] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 387. | Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230-2237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 388. | Wang L, Ou JJ. Regulation of Autophagy by Hepatitis C Virus for Its Replication. DNA Cell Biol. 2018;37:287-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 389. | Wang L, Kim JY, Liu HM, Lai MMC, Ou JJ. HCV-induced autophagosomes are generated via homotypic fusion of phagophores that mediate HCV RNA replication. PLoS Pathog. 2017;13:e1006609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 390. | Shrivastava S, Devhare P, Sujijantarat N, Steele R, Kwon YC, Ray R, Ray RB. Knockdown of Autophagy Inhibits Infectious Hepatitis C Virus Release by the Exosomal Pathway. J Virol. 2016;90:1387-1396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 391. | Mori H, Fukuhara T, Ono C, Tamura T, Sato A, Fauzyah Y, Wada M, Okamoto T, Noda T, Yoshimori T, Matsuura Y. Induction of selective autophagy in cells replicating hepatitis C virus genome. J Gen Virol. 2018;99:1643-1657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 392. | Mohl BP, Tedbury PR, Griffin S, Harris M. Hepatitis C virus-induced autophagy is independent of the unfolded protein response. J Virol. 2012;86:10724-10732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 393. | Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046-14051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 379] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 394. | Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 374] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 395. | Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013;87:10612-10627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 396. | Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37-56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 269] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 397. | Chan ST, Lee J, Narula M, Ou JJ. Suppression of Host Innate Immune Response by Hepatitis C Virus via Induction of Autophagic Degradation of TRAF6. J Virol. 2016;90:10928-10935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 398. | Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054-1061. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 286] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 399. | Dash S, Chava S, Aydin Y, Chandra PK, Ferraris P, Chen W, Balart LA, Wu T, Garry RF. Hepatitis C Virus Infection Induces Autophagy as a Prosurvival Mechanism to Alleviate Hepatic ER-Stress Response. Viruses. 2016;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 400. | Taguwa S, Kambara H, Fujita N, Noda T, Yoshimori T, Koike K, Moriishi K, Matsuura Y. Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J Virol. 2011;85:13185-13194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 401. | Vescovo T, Romagnoli A, Perdomo AB, Corazzari M, Ciccosanti F, Alonzi T, Nardacci R, Ippolito G, Tripodi M, Garcia-Monzon C, Lo Iacono O, Piacentini M, Fimia GM. Autophagy protects cells from HCV-induced defects in lipid metabolism. Gastroenterology 2012; 142: 644-653. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 402. | Chu VC, Bhattacharya S, Nomoto A, Lin J, Zaidi SK, Oberley TD, Weinman SA, Azhar S, Huang TT. Persistent expression of hepatitis C virus non-structural proteins leads to increased autophagy and mitochondrial injury in human hepatoma cells. PLoS One. 2011;6:e28551. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 403. | Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci USA. 2014;111:6413-6418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 210] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 404. | Jassey A, Liu CH, Changou CA, Richardson CD, Hsu HY, Lin LT. Hepatitis C Virus Non-Structural Protein 5A (NS5A) Disrupts Mitochondrial Dynamics and Induces Mitophagy. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 405. | Hara Y, Yanatori I, Ikeda M, Kiyokage E, Nishina S, Tomiyama Y, Toida K, Kishi F, Kato N, Imamura M, Chayama K, Hino K. Hepatitis C virus core protein suppresses mitophagy by interacting with parkin in the context of mitochondrial depolarization. Am J Pathol. 2014;184:3026-3039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 406. | Ren H, Elgner F, Jiang B, Himmelsbach K, Medvedev R, Ploen D, Hildt E. The Autophagosomal SNARE Protein Syntaxin 17 Is an Essential Factor for the Hepatitis C Virus Life Cycle. J Virol. 2016;90:5989-6000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 407. | Kurt R, Chandra PK, Aboulnasr F, Panigrahi R, Ferraris P, Aydin Y, Reiss K, Wu T, Balart LA, Dash S. Chaperone-Mediated Autophagy Targets IFNAR1 for Lysosomal Degradation in Free Fatty Acid Treated HCV Cell Culture. PLoS One. 2015;10:e0125962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 408. | Matsui C, Deng L, Minami N, Abe T, Koike K, Shoji I. Hepatitis C Virus NS5A Protein Promotes the Lysosomal Degradation of Hepatocyte Nuclear Factor 1α via Chaperone-Mediated Autophagy. J Virol. 2018;92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 409. | Zhang MQ, Li JR, Peng ZG, Zhang JP. Differential Effects of Autophagy-Related 10 Protein on HCV Replication and Autophagy Flux Are Mediated by Its Cysteine44 and Cysteine135. Front Immunol. 2018;9:2176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 410. | Zhao Q, Hu ZY, Zhang JP, Jiang JD, Ma YY, Li JR, Peng ZG, Chen JH. Dual Roles of Two Isoforms of Autophagy-related Gene ATG10 in HCV-Subgenomic replicon Mediated Autophagy Flux and Innate Immunity. Sci Rep. 2017;7:11250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 411. | Mizui T, Yamashina S, Tanida I, Takei Y, Ueno T, Sakamoto N, Ikejima K, Kitamura T, Enomoto N, Sakai T, Kominami E, Watanabe S. Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J Gastroenterol. 2010;45:195-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 412. | Kim JY, Ou JJ. Regulation of Apolipoprotein E Trafficking by Hepatitis C Virus-Induced Autophagy. J Virol. 2018;92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 413. | Zhang L. Autophagy in hepatitis B or C virus infection: An incubator and a potential therapeutic target. Life Sci. 2020;242:117206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 414. | Lazar C, Uta M, Branza-Nichita N. Modulation of the unfolded protein response by the human hepatitis B virus. Front Microbiol. 2014;5:433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 415. | Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241-2249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 416. | Rautou PE, Cazals-Hatem D, Feldmann G, Mansouri A, Grodet A, Barge S, Martinot-Peignoux M, Duces A, Bièche I, Lebrec D, Bedossa P, Paradis V, Marcellin P, Valla D, Asselah T, Moreau R. Changes in autophagic response in patients with chronic hepatitis C virus infection. Am J Pathol. 2011;178:2708-2715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 417. | Aweya JJ, Mak TM, Lim SG, Tan YJ. The p7 protein of the hepatitis C virus induces cell death differently from the influenza A virus viroporin M2. Virus Res. 2013;172:24-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 418. | Ríos-Ocampo WA, Daemen T, Buist-Homan M, Faber KN, Navas MC, Moshage H. Hepatitis C virus core or NS3/4A protein expression preconditions hepatocytes against oxidative stress and endoplasmic reticulum stress. Redox Rep. 2019;24:17-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 419. | Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol. 2011;85:10561-10571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 420. | Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 421. | Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1221] [Cited by in F6Publishing: 1701] [Article Influence: 243.0] [Reference Citation Analysis (0)] |
| 422. | Williams MJ, Clouston AD, Forbes SJ. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology. 2014;146:349-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 214] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 423. | Sato K, Marzioni M, Meng F, Francis H, Glaser S, Alpini G. Ductular Reaction in Liver Diseases: Pathological Mechanisms and Translational Significances. Hepatology. 2019;69:420-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 244] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 424. | Gouw AS, Clouston AD, Theise ND. Ductular reactions in human liver: diversity at the interface. Hepatology. 2011;54:1853-1863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 425. | Clouston AD, Powell EE, Walsh MJ, Richardson MM, Demetris AJ, Jonsson JR. Fibrosis correlates with a ductular reaction in hepatitis C: roles of impaired replication, progenitor cells and steatosis. Hepatology. 2005;41:809-818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 257] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 426. | Richardson MM, Jonsson JR, Powell EE, Brunt EM, Neuschwander-Tetri BA, Bhathal PS, Dixon JB, Weltman MD, Tilg H, Moschen AR, Purdie DM, Demetris AJ, Clouston AD. Progressive fibrosis in nonalcoholic steatohepatitis: association with altered regeneration and a ductular reaction. Gastroenterology. 2007;133:80-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 364] [Cited by in F6Publishing: 332] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 427. | Wood MJ, Gadd VL, Powell LW, Ramm GA, Clouston AD. Ductular reaction in hereditary hemochromatosis: the link between hepatocyte senescence and fibrosis progression. Hepatology. 2014;59:848-857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 428. | Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 277] [Cited by in F6Publishing: 276] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 429. | Song Y, Zhao Y, Wang F, Tao L, Xiao J, Yang C. Autophagy in hepatic fibrosis. Biomed Res Int. 2014;2014:436242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 430. | Kwanten WJ, Martinet W, Michielsen PP, Francque SM. Role of autophagy in the pathophysiology of nonalcoholic fatty liver disease: a controversial issue. World J Gastroenterol. 2014;20:7325-7338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 75] [Cited by in F6Publishing: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 431. | Li J, Chen K, Li S, Feng J, Liu T, Wang F, Zhang R, Xu S, Zhou Y, Zhou S, Xia Y, Lu J, Zhou Y, Guo C. Protective effect of fucoidan from Fucus vesiculosus on liver fibrosis via the TGF-β1/Smad pathway-mediated inhibition of extracellular matrix and autophagy. Drug Des Devel Ther. 2016;10:619-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 432. | Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3:17-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 223] [Cited by in F6Publishing: 260] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 433. | Mao YQ, Fan XM. Autophagy: A new therapeutic target for liver fibrosis. World J Hepatol. 2015;7:1982-1986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 434. | Miyamae Y, Nishito Y, Nakai N, Nagumo Y, Usui T, Masuda S, Kambe T, Nagao M. Tetrandrine induces lipid accumulation through blockade of autophagy in a hepatic stellate cell line. Biochem Biophys Res Commun. 2016;477:40-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 435. | He W, Wang B, Yang J, Zhuang Y, Wang L, Huang X, Chen J. Chloroquine improved carbon tetrachloride-induced liver fibrosis through its inhibition of the activation of hepatic stellate cells: role of autophagy. Biol Pharm Bull. 2014;37:1505-1509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 436. | Lee AY, Lee JW, Kim JE, Mock HJ, Park S, Kim S, Hong SH, Kim JY, Park EJ, Kang KS, Kim KP, Cho MH. Dihydroceramide is a key metabolite that regulates autophagy and promotes fibrosis in hepatic steatosis model. Biochem Biophys Res Commun. 2017;494:460-469. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 437. | Zhang XW, Mi S, Li Z, Zhou JC, Xie J, Hua F, Li K, Cui B, Lv XX, Yu JJ, Hu ZW. Antagonism of Interleukin-17A ameliorates experimental hepatic fibrosis by restoring the IL-10/STAT3-suppressed autophagy in hepatocytes. Oncotarget. 2017;8:9922-9934. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 438. | Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 352] [Cited by in F6Publishing: 330] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 439. | Hernández-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 495] [Cited by in F6Publishing: 485] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 440. | Thoen LF, Guimarães EL, Dollé L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353-1360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 291] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 441. | Thoen LF, Guimarães EL, Grunsven LA. Autophagy: a new player in hepatic stellate cell activation. Autophagy. 2012;8:126-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 442. | Hernández-Gea V, Hilscher M, Rozenfeld R, Lim MP, Nieto N, Werner S, Devi LA, Friedman SL. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol. 2013;59:98-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 176] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 443. | Vere CC, Streba CT, Streba L, Rogoveanu I. Lipid serum profile in patients with viral liver cirrhosis. Med Princ Pract. 2012;21:566-568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 444. | Kim KM, Han CY, Kim JY, Cho SS, Kim YS, Koo JH, Lee JM, Lim SC, Kang KW, Kim JS, Hwang SJ, Ki SH, Kim SG. Gα12 overexpression induced by miR-16 dysregulation contributes to liver fibrosis by promoting autophagy in hepatic stellate cells. J Hepatol. 2018;68:493-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 445. | Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M, Lu L. Exosomes derived from miR-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med. 2017;21:2491-2502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 304] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 446. | Deng J, Huang Q, Wang Y, Shen P, Guan F, Li J, Huang H, Shi C. Hypoxia-inducible factor-1alpha regulates autophagy to activate hepatic stellate cells. Biochem Biophys Res Commun. 2014;454:328-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 447. | Fu MY, He YJ, Lv X, Liu ZH, Shen Y, Ye GR, Deng YM, Shu JC. Transforming growth factorβ1 reduces apoptosis via autophagy activation in hepatic stellate cells. Mol Med Rep. 2014;10:1282-1288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 448. | Li J, Zeng C, Zheng B, Liu C, Tang M, Jiang Y, Chang Y, Song W, Wang Y, Yang C. HMGB1-induced autophagy facilitates hepatic stellate cells activation: a new pathway in liver fibrosis. Clin Sci (Lond). 2018;132:1645-1667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 449. | Zhang Z, Guo M, Zhao S, Shao J, Zheng S. ROS-JNK1/2-dependent activation of autophagy is required for the induction of anti-inflammatory effect of dihydroartemisinin in liver fibrosis. Free Radic Biol Med. 2016;101:272-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 450. | Kim RS, Hasegawa D, Goossens N, Tsuchida T, Athwal V, Sun X, Robinson CL, Bhattacharya D, Chou HI, Zhang DY, Fuchs BC, Lee Y, Hoshida Y, Friedman SL. The XBP1 Arm of the Unfolded Protein Response Induces Fibrogenic Activity in Hepatic Stellate Cells Through Autophagy. Sci Rep. 2016;6:39342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 451. | Hong Y, Li S, Wang J, Li Y. In vitro inhibition of hepatic stellate cell activation by the autophagy-related lipid droplet protein ATG2A. Sci Rep. 2018;8:9232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 452. | Wu L, Zhang Q, Mo W, Feng J, Li S, Li J, Liu T, Xu S, Wang W, Lu X, Yu Q, Chen K, Xia Y, Lu J, Xu L, Zhou Y, Fan X, Guo C. Quercetin prevents hepatic fibrosis by inhibiting hepatic stellate cell activation and reducing autophagy via the TGF-β1/Smads and PI3K/Akt pathways. Sci Rep. 2017;7:9289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 162] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 453. | Qiu YN, Wang GH, Zhou F, Hao JJ, Tian L, Guan LF, Geng XK, Ding YC, Wu HW, Zhang KZ. PM2.5 induces liver fibrosis via triggering ROS-mediated mitophagy. Ecotoxicol Environ Saf. 2019;167:178-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 454. | Tian Z, Chen Y, Yao N, Hu C, Wu Y, Guo D, Liu J, Yang Y, Chen T, Zhao Y, He Y. Role of mitophagy regulation by ROS in hepatic stellate cells during acute liver failure. Am J Physiol Gastrointest Liver Physiol. 2018;315:G374-G384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 455. | Li J, Zhao YR, Tian Z. Roles of hepatic stellate cells in acute liver failure: From the perspective of inflammation and fibrosis. World J Hepatol. 2019;11:412-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 456. | Wu H, Chen G, Wang J, Deng M, Yuan F, Gong J. TIM-4 interference in Kupffer cells against CCL4-induced liver fibrosis by mediating Akt1/Mitophagy signalling pathway. Cell Prolif. 2020;53:e12731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 457. | Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, Lau SL, Atkins AR, Barish GD, Gunton JE, Liddle C, Downes M, Evans RM. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601-613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 488] [Cited by in F6Publishing: 488] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 458. | Duran A, Hernandez ED, Reina-Campos M, Castilla EA, Subramaniam S, Raghunandan S, Roberts LR, Kisseleva T, Karin M, Diaz-Meco MT, Moscat J. p62/SQSTM1 by Binding to Vitamin D Receptor Inhibits Hepatic Stellate Cell Activity, Fibrosis, and Liver Cancer. Cancer Cell. 2016;30:595-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 166] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 459. | Zhang Z, Yao Z, Wang L, Ding H, Shao J, Chen A, Zhang F, Zheng S. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14:2083-2103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 294] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 460. | Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 621] [Cited by in F6Publishing: 853] [Article Influence: 121.9] [Reference Citation Analysis (0)] |
| 461. | Lodder J, Denaës T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM, Lotersztajn S, Teixeira-Clerc F. Macrophage autophagy protects against liver fibrosis in mice. Autophagy. 2015;11:1280-1292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 190] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 462. | Ilyas G, Zhao E, Liu K, Lin Y, Tesfa L, Tanaka KE, Czaja MJ. Macrophage autophagy limits acute toxic liver injury in mice through down regulation of interleukin-1β. J Hepatol. 2016;64:118-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 463. | Sun K, Xu L, Jing Y, Han Z, Chen X, Cai C, Zhao P, Zhao X, Yang L, Wei L. Autophagy-deficient Kupffer cells promote tumorigenesis by enhancing mtROS-NF-κB-IL1α/β-dependent inflammation and fibrosis during the preneoplastic stage of hepatocarcinogenesis. Cancer Lett. 2017;388:198-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 464. | Habib A, Chokr D, Wan J, Hegde P, Mabire M, Siebert M, Ribeiro-Parenti L, Le Gall M, Lettéron P, Pilard N, Mansouri A, Brouillet A, Tardelli M, Weiss E, Le Faouder P, Guillou H, Cravatt BF, Moreau R, Trauner M, Lotersztajn S. Inhibition of monoacylglycerol lipase, an anti-inflammatory and antifibrogenic strategy in the liver. Gut. 2019;68:522-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 465. | Ruart M, Chavarria L, Campreciós G, Suárez-Herrera N, Montironi C, Guixé-Muntet S, Bosch J, Friedman SL, Garcia-Pagán JC, Hernández-Gea V. Impaired endothelial autophagy promotes liver fibrosis by aggravating the oxidative stress response during acute liver injury. J Hepatol. 2019;70:458-469. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 466. | Luo X, Wang D, Zhu X, Wang G, You Y, Ning Z, Li Y, Jin S, Huang Y, Hu Y, Chen T, Meng Y, Li X. Autophagic degradation of caveolin-1 promotes liver sinusoidal endothelial cells defenestration. Cell Death Dis. 2018;9:576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 467. | Boteon YL, Laing R, Mergental H, Reynolds GM, Mirza DF, Afford SC, Bhogal RH. Mechanisms of autophagy activation in endothelial cell and their targeting during normothermic machine liver perfusion. World J Gastroenterol. 2017;23:8443-8451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 468. | Pozniak KN, Pearen MA, Pereira TN, Kramer CSM, Kalita-De Croft P, Nawaratna SK, Fernandez-Rojo MA, Gobert GN, Tirnitz-Parker JEE, Olynyk JK, Shepherd RW, Lewindon PJ, Ramm GA. Taurocholate Induces Biliary Differentiation of Liver Progenitor Cells Causing Hepatic Stellate Cell Chemotaxis in the Ductular Reaction: Role in Pediatric Cystic Fibrosis Liver Disease. Am J Pathol. 2017;187:2744-2757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 469. | Hung TM, Huang YJ, Lin YC, Chen YH, Wu YM, Lee PH. A critical role of autophagy in regulating the mesenchymal transition of ductular cells in liver cirrhosis. Sci Rep. 2019;9:10673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 470. | Hung TM, Hsiao CC, Lin CW, Lee PH. Complex Cell Type-Specific Roles of Autophagy in Liver Fibrosis and Cirrhosis. Pathogens. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 471. | Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1454] [Cited by in F6Publishing: 1549] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 472. | Yazdani HO, Huang H, Tsung A. Autophagy: Dual Response in the Development of Hepatocellular Carcinoma. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 473. | Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 921] [Cited by in F6Publishing: 1008] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 474. | Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 324] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 475. | Lim SC, Han SI. Ursodeoxycholic acid effectively kills drug-resistant gastric cancer cells through induction of autophagic death. Oncol Rep. 2015;34:1261-1268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 476. | White DL, Thrift AP, Kanwal F, Davila J, El-Serag HB. Incidence of Hepatocellular Carcinoma in All 50 United States, From 2000 Through 2012. Gastroenterology 2017; 152: 812-820. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 313] [Cited by in F6Publishing: 303] [Article Influence: 43.3] [Reference Citation Analysis (1)] |
| 477. | West J, Card TR, Aithal GP, Fleming KM. Risk of hepatocellular carcinoma among individuals with different aetiologies of cirrhosis: a population-based cohort study. Aliment Pharmacol Ther. 2017;45:983-990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 478. | Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 2012; 143: 177-87. e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 479. | Ni HM, Chao X, Yang H, Deng F, Wang S, Bai Q, Qian H, Cui Y, Cui W, Shi Y, Zong WX, Wang Z, Yang L, Ding WX. Dual Roles of Mammalian Target of Rapamycin in Regulating Liver Injury and Tumorigenesis in Autophagy-Defective Mouse Liver. Hepatology. 2019;70:2142-2155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 480. | Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta. 2013;1836:15-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 481. | Sun K, Guo XL, Zhao QD, Jing YY, Kou XR, Xie XQ, Zhou Y, Cai N, Gao L, Zhao X, Zhang SS, Song JR, Li D, Deng WJ, Li R, Wu MC, Wei LX. Paradoxical role of autophagy in the dysplastic and tumor-forming stages of hepatocarcinoma development in rats. Cell Death Dis. 2013;4:e501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 482. | Taniguchi K, Yamachika S, He F, Karin M. p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett. 2016;590:2375-2397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 483. | Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res. 2012;18:370-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 237] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 484. | Wu DH, Jia CC, Chen J, Lin ZX, Ruan DY, Li X, Lin Q, Min-Dong, Ma XK, Wan XB, Cheng N, Chen ZH, Xing YF, Wu XY, Wen JY. Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol. 2014;35:12225-12233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 485. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 923] [Cited by in F6Publishing: 1008] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 486. | Bao L, Chandra PK, Moroz K, Zhang X, Thung SN, Wu T, Dash S. Impaired autophagy response in human hepatocellular carcinoma. Exp Mol Pathol. 2014;96:149-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 487. | Shi YH, Ding ZB, Zhou J, Qiu SJ, Fan J. Prognostic significance of Beclin 1-dependent apoptotic activity in hepatocellular carcinoma. Autophagy. 2009;5:380-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 488. | Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, Fan J. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167-9175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 223] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 489. | DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1492] [Cited by in F6Publishing: 1670] [Article Influence: 128.5] [Reference Citation Analysis (0)] |
| 490. | Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576-22591. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 942] [Cited by in F6Publishing: 1125] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 491. | Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275-3285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 586] [Cited by in F6Publishing: 665] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 492. | Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275-284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 430] [Cited by in F6Publishing: 460] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 493. | Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, Jaeschke H, Ding WX. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 200] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 494. | Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, Ohishi M, Endo K, Uemura T, Nishito Y, Okuda S, Obata M, Kouno T, Imamura R, Tada Y, Obata R, Yasuda D, Takahashi K, Fujimura T, Pi J, Lee MS, Ueno T, Ohe T, Mashino T, Wakai T, Kojima H, Okabe T, Nagano T, Motohashi H, Waguri S, Soga T, Yamamoto M, Tanaka K, Komatsu M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 247] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 495. | Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 947] [Cited by in F6Publishing: 1038] [Article Influence: 86.5] [Reference Citation Analysis (0)] |
| 496. | Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, Raschioni C, Testore E, Roncalli M, Giordano S, Columbano A. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61:1088-1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 497. | Turcios L, Chacon E, Garcia C, Eman P, Cornea V, Jiang J, Spear B, Liu C, Watt DS, Marti F, Gedaly R. Autophagic flux modulation by Wnt/β-catenin pathway inhibition in hepatocellular carcinoma. PLoS One. 2019;14:e0212538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 498. | Wang W, Xu L, Liu P, Jairam K, Yin Y, Chen K, Sprengers D, Peppelenbosch MP, Pan Q, Smits R. Blocking Wnt Secretion Reduces Growth of Hepatocellular Carcinoma Cell Lines Mostly Independent of β-Catenin Signaling. Neoplasia. 2016;18:711-723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 499. | Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, Sugimoto K, Miyazono K. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69:8844-8852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 500. | Xie R, Wang F, McKeehan WL, Liu L. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis. Cancer Res. 2011;71:7537-7546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 501. | Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Chang YG, Kim MG, Park WS, Lee JY, Lee SY, Chu IS, Nam SW. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56:644-657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 502. | Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Lee JY, Park H, Nam SW. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7:e34265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 503. | Zou M, Lu N, Hu C, Liu W, Sun Y, Wang X, You Q, Gu C, Xi T, Guo Q. Beclin 1-mediated autophagy in hepatocellular carcinoma cells: implication in anticancer efficiency of oroxylin A via inhibition of mTOR signaling. Cell Signal. 2012;24:1722-1732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 504. | Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360-3371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 505. | Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y, Liu J, Jia L. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 2013;20:235-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 506. | Wang Y, Han C, Lu L, Magliato S, Wu T. Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology. 2013;58:995-1010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 507. | Chang CP, Yang MC, Liu HS, Lin YS, Lei HY. Concanavalin A induces autophagy in hepatoma cells and has a therapeutic effect in a murine in situ hepatoma model. Hepatology. 2007;45:286-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 142] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 508. | Lei HY, Chang CP. Induction of autophagy by concanavalin A and its application in anti-tumor therapy. Autophagy. 2007;3:402-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 509. | Chang CP, Yang MC, Lei HY. Concanavalin A/IFN-gamma triggers autophagy-related necrotic hepatocyte death through IRGM1-mediated lysosomal membrane disruption. PLoS One. 2011;6:e28323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 510. | Qian H, Yang Y. Alterations of cellular organelles in human liver-derived hepatoma G2 cells induced by adriamycin. Anticancer Drugs. 2009;20:779-786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 511. | Yang F, Gao YH, Wu KW, Deng R, Li DD, Wei ZX, Jiang S, Wu XQ, Feng GK, Li HJ, Zhu XF. A novel sesquiterpene Hirsutanol A induces autophagical cell death in human hepatocellular carcinoma cells by increasing reactive oxygen species. Chin J Cancer. 2010;29:655-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 512. | Xie SQ, Li Q, Zhang YH, Wang JH, Mei ZH, Zhao J, Wang CJ. NPC-16, a novel naphthalimide-polyamine conjugate, induced apoptosis and autophagy in human hepatoma HepG2 cells and Bel-7402 cells. Apoptosis. 2011;16:27-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 513. | Zhang JQ, Li YM, Liu T, He WT, Chen YT, Chen XH, Li X, Zhou WC, Yi JF, Ren ZJ. Antitumor effect of matrine in human hepatoma G2 cells by inducing apoptosis and autophagy. World J Gastroenterol. 2010;16:4281-4290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 97] [Cited by in F6Publishing: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 514. | Wang N, Feng Y, Zhu M, Tsang CM, Man K, Tong Y, Tsao SW. Berberine induces autophagic cell death and mitochondrial apoptosis in liver cancer cells: the cellular mechanism. J Cell Biochem. 2010;111:1426-1436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 515. | Wang N, Pan W, Zhu M, Zhang M, Hao X, Liang G, Feng Y. Fangchinoline induces autophagic cell death via p53/sestrin2/AMPK signalling in human hepatocellular carcinoma cells. Br J Pharmacol. 2011;164:731-742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 516. | Hou Q, Tang X, Liu H, Tang J, Yang Y, Jing X, Xiao Q, Wang W, Gou X, Wang Z. Berberine induces cell death in human hepatoma cells in vitro by downregulating CD147. Cancer Sci. 2011;102:1287-1292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 517. | Qian H, Yang Y, Wang X. Curcumin enhanced adriamycin-induced human liver-derived Hepatoma G2 cell death through activation of mitochondria-mediated apoptosis and autophagy. Eur J Pharm Sci. 2011;43:125-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 518. | Andrzejak M, Price M, Kessel DH. Apoptotic and autophagic responses to photodynamic therapy in 1c1c7 murine hepatoma cells. Autophagy. 2011;7:979-984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 519. | Hu M, Huang H, Zhao R, Li P, Li M, Miao H, Chen N, Chen M. AZD8055 induces cell death associated with autophagy and activation of AMPK in hepatocellular carcinoma. Oncol Rep. 2014;31:649-656. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 520. | Song J, Qu Z, Guo X, Zhao Q, Zhao X, Gao L, Sun K, Shen F, Wu M, Wei L. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 521. | Chen LH, Loong CC, Su TL, Lee YJ, Chu PM, Tsai ML, Tsai PH, Tu PH, Chi CW, Lee HC, Chiou SH. Autophagy inhibition enhances apoptosis triggered by BO-1051, an N-mustard derivative, and involves the ATM signaling pathway. Biochem Pharmacol. 2011;81:594-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 522. | Ding ZB, Hui B, Shi YH, Zhou J, Peng YF, Gu CY, Yang H, Shi GM, Ke AW, Wang XY, Song K, Dai Z, Shen YH, Fan J. Autophagy activation in hepatocellular carcinoma contributes to the tolerance of oxaliplatin via reactive oxygen species modulation. Clin Cancer Res. 2011;17:6229-6238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 523. | Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, Hosui A, Ishida H, Tatsumi T, Kanto T, Hiramatsu N, Fujita N, Yoshimori T, Hayashi N. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548-557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 207] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 524. | Du H, Yang W, Chen L, Shi M, Seewoo V, Wang J, Lin A, Liu Z, Qiu W. Role of autophagy in resistance to oxaliplatin in hepatocellular carcinoma cells. Oncol Rep. 2012;27:143-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 525. | Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F, Xia Q. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423:826-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 526. | Yu HC, Lin CS, Tai WT, Liu CY, Shiau CW, Chen KF. Nilotinib induces autophagy in hepatocellular carcinoma through AMPK activation. J Biol Chem. 2013;288:18249-18259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 527. | Wu B, Cui J, Yang XM, Liu ZY, Song F, Li L, Jiang JL, Chen ZN. Cytoplasmic fragment of CD147 generated by regulated intramembrane proteolysis contributes to HCC by promoting autophagy. Cell Death Dis. 2017;8:e2925. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 528. | Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW, Wang XY, Dai Z, Peng YF, Gu CY, Qiu SJ, Fan J. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159-1172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 259] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 529. | Bareford MD, Park MA, Yacoub A, Hamed HA, Tang Y, Cruickshanks N, Eulitt P, Hubbard N, Tye G, Burow ME, Fisher PB, Moran RG, Nephew KP, Grant S, Dent P. Sorafenib enhances pemetrexed cytotoxicity through an autophagy-dependent mechanism in cancer cells. Cancer Res. 2011;71:4955-4967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 530. | Li X, Zhou Y, Yang L, Ma Y, Peng X, Yang S, Li H, Liu J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. 2020;235:3402-3413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 531. | Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K, Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 155] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 532. | Prieto-Domínguez N, Ordóñez R, Fernández A, Méndez-Blanco C, Baulies A, Garcia-Ruiz C, Fernández-Checa JC, Mauriz JL, González-Gallego J. Melatonin-induced increase in sensitivity of human hepatocellular carcinoma cells to sorafenib is associated with reactive oxygen species production and mitophagy. J Pineal Res. 2016;61:396-407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 533. | Li W, Li Y, Siraj S, Jin H, Fan Y, Yang X, Huang X, Wang X, Wang J, Liu L, Du L, Chen Q. FUN14 Domain-Containing 1-Mediated Mitophagy Suppresses Hepatocarcinogenesis by Inhibition of Inflammasome Activation in Mice. Hepatology. 2019;69:604-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 534. | Huang Q, Zhan L, Cao H, Li J, Lyu Y, Guo X, Zhang J, Ji L, Ren T, An J, Liu B, Nie Y, Xing J. Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways. Autophagy. 2016;12:999-1014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 247] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 535. | Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan ST, Cho C, Machida K, Chen D, Ou JJ. Mitophagy Controls the Activities of Tumor Suppressor p53 to Regulate Hepatic Cancer Stem Cells. Mol Cell 2017; 68: 281-292. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 536. | Gómez de Cedrón M, Ramírez de Molina A. Microtargeting cancer metabolism: opening new therapeutic windows based on lipid metabolism. J Lipid Res. 2016;57:193-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 537. | Zhao T, Du H, Ding X, Walls K, Yan C. Activation of mTOR pathway in myeloid-derived suppressor cells stimulates cancer cell proliferation and metastasis in lal(-/-) mice. Oncogene. 2015;34:1938-1948. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 538. | Mukhopadhyay S, Schlaepfer IR, Bergman BC, Panda PK, Praharaj PP, Naik PP, Agarwal R, Bhutia SK. ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radic Biol Med. 2017;104:199-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 539. | Lu GD, Ang YH, Zhou J, Tamilarasi J, Yan B, Lim YC, Srivastava S, Salto-Tellez M, Hui KM, Shen HM, Nguyen LN, Tan BC, Silver DL, Hooi SC. CCAAT/enhancer binding protein α predicts poorer prognosis and prevents energy starvation-induced cell death in hepatocellular carcinoma. Hepatology. 2015;61:965-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 540. | Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, Cuervo AM. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3:109ra117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 190] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 541. | Chava S, Lee C, Aydin Y, Chandra PK, Dash A, Chedid M, Thung SN, Moroz K, Wu T, Nayak NC, Dash S. Chaperone-mediated autophagy compensates for impaired macroautophagy in the cirrhotic liver to promote hepatocellular carcinoma. Oncotarget. 2017;8:40019-40036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 542. | Dash S, Aydin Y, Moroz K. Chaperone-Mediated Autophagy in the Liver: Good or Bad? Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 543. | Wu WKK, Zhang L, Chan MTV. Autophagy, NAFLD and NAFLD-Related HCC. Adv Exp Med Biol. 2018;1061:127-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 544. | Niture S, Gyamfi MA, Lin M, Chimeh U, Dong X, Zheng W, Moore J, Kumar D. TNFAIP8 regulates autophagy, cell steatosis, and promotes hepatocellular carcinoma cell proliferation. Cell Death Dis. 2020;11:178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 545. | Liu L, Liao JZ, He XX, Li PY. The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget. 2017;8:57707-57722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 546. | Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4643-4651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 119] [Cited by in F6Publishing: 140] [Article Influence: 23.3] [Reference Citation Analysis (2)] |
| 547. | Yang S, Yang L, Li X, Li B, Li Y, Zhang X, Ma Y, Peng X, Jin H, Li H. New insights into autophagy in hepatocellular carcinoma: mechanisms and therapeutic strategies. Am J Cancer Res. 2019;9:1329-1353. [PubMed] [Cited in This Article: ] |
| 548. | Chen P, Bonaldo P. Role of macrophage polarization in tumor angiogenesis and vessel normalization: implications for new anticancer therapies. Int Rev Cell Mol Biol. 2013;301:1-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 549. | Lin H, Yan J, Wang Z, Hua F, Yu J, Sun W, Li K, Liu H, Yang H, Lv Q, Xue J, Hu ZW. Loss of immunity-supported senescence enhances susceptibility to hepatocellular carcinogenesis and progression in Toll-like receptor 2-deficient mice. Hepatology. 2013;57:171-182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 550. | Chen W, Ma T, Shen XN, Xia XF, Xu GD, Bai XL, Liang TB. Macrophage-induced tumor angiogenesis is regulated by the TSC2-mTOR pathway. Cancer Res. 2012;72:1363-1372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 551. | Chen KD, Lin CC, Tsai MC, Huang KT, Chiu KW. Tumor microenvironment mediated by suppression of autophagic flux drives liver malignancy. Biomed J. 2018;41:163-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 552. | Hu T, Li P, Luo Z, Chen X, Zhang J, Wang C, Chen P, Dong Z. Chloroquine inhibits hepatocellular carcinoma cell growth in vitro and in vivo. Oncol Rep. 2016;35:43-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 553. | Wang Y, Zhao H, Wang D, Hao M, Kong C, Zhao X, Gao Y, Li J, Liu B, Yang B, Zhang H, Jiang J. Inhibition of Autophagy Promoted Apoptosis and Suppressed Growth of Hepatocellular Carcinoma Upon Photothermal Exposure. J Biomed Nanotechnol. 2019;15:813-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 554. | Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654-666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 685] [Cited by in F6Publishing: 693] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 555. | Di Fazio P, Matrood S. Targeting autophagy in liver cancer. Transl Gastroenterol Hepatol. 2018;3:39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 556. | Peng YF, Shi YH, Ding ZB, Ke AW, Gu CY, Hui B, Zhou J, Qiu SJ, Dai Z, Fan J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9:2056-2068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 178] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 557. | Gao L, Song JR, Zhang JW, Zhao X, Zhao QD, Sun K, Deng WJ, Li R, Lv G, Cheng HY, Wei LX. Chloroquine promotes the anticancer effect of TACE in a rabbit VX2 Liver tumor model. Int J Biol Sci. 2013;9:322-330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 558. | Wang X, Deng Q, Feng K, Chen S, Jiang J, Xia F, Ma K, Bie P. Insufficient radiofrequency ablation promotes hepatocellular carcinoma cell progression via autophagy and the CD133 feedback loop. Oncol Rep. 2018;40:241-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 559. | Zhao Z, Wu J, Liu X, Liang M, Zhou X, Ouyang S, Yao J, Wang J, Luo B. Insufficient radiofrequency ablation promotes proliferation of residual hepatocellular carcinoma via autophagy. Cancer Lett. 2018;421:73-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 560. | Prieto-Domínguez N, Ordóñez R, Fernández A, García-Palomo A, Muntané J, González-Gallego J, Mauriz JL. Modulation of Autophagy by Sorafenib: Effects on Treatment Response. Front Pharmacol. 2016;7:151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 561. | Zhao P, Li M, Wang Y, Chen Y, He C, Zhang X, Yang T, Lu Y, You J, Lee RJ, Xiang G. Enhancing anti-tumor efficiency in hepatocellular carcinoma through the autophagy inhibition by miR-375/sorafenib in lipid-coated calcium carbonate nanoparticles. Acta Biomater. 2018;72:248-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 562. | Hou YJ, Dong LW, Tan YX, Yang GZ, Pan YF, Li Z, Tang L, Wang M, Wang Q, Wang HY. Inhibition of active autophagy induces apoptosis and increases chemosensitivity in cholangiocarcinoma. Lab Invest. 2011;91:1146-1157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 563. | Sasaki M, Nitta T, Sato Y, Nakanuma Y. Autophagy may occur at an early stage of cholangiocarcinogenesis via biliary intraepithelial neoplasia. Hum Pathol. 2015;46:202-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 564. | Dong LW, Hou YJ, Tan YX, Tang L, Pan YF, Wang M, Wang HY. Prognostic significance of Beclin 1 in intrahepatic cholangiocellular carcinoma. Autophagy. 2011;7:1222-1229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 565. | Wang TT, Cao QH, Chen MY, Xia Q, Fan XJ, Ma XK, Lin Q, Jia CC, Dong M, Ruan DY, Lin ZX, Wen JY, Wei L, Li X, Chen ZH, Wang L, Wu XY, Wan XB. Beclin 1 deficiency correlated with lymph node metastasis, predicts a distinct outcome in intrahepatic and extrahepatic cholangiocarcinoma. PLoS One. 2013;8:e80317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 566. | Qu X, Sheng J, Shen L, Su J, Xu Y, Xie Q, Wu Y, Zhang X, Sun L. Autophagy inhibitor chloroquine increases sensitivity to cisplatin in QBC939 cholangiocarcinoma cells by mitochondrial ROS. PLoS One. 2017;12:e0173712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 567. | Jia B, Xue Y, Yan X, Li J, Wu Y, Guo R, Zhang J, Zhang L, Li Y, Liu Y, Sun L. Autophagy inhibitor chloroquine induces apoptosis of cholangiocarcinoma cells via endoplasmic reticulum stress. Oncol Lett. 2018;16:3509-3516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 568. | O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM, Zhu AX, Land H, Bardeesy N, Hezel AF. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557-1567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 157] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 569. | Huang JL, Hezel AF. Autophagy in intra-hepatic cholangiocarcinoma. Autophagy. 2012;8:1148-1149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 570. | Hong ZF, Zhao WX, Yin ZY, Xie CR, Xu YP, Chi XQ, Zhang S, Wang XM. Capsaicin Enhances the Drug Sensitivity of Cholangiocarcinoma through the Inhibition of Chemotherapeutic-Induced Autophagy. PLoS One. 2015;10:e0121538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 571. | Ding X, Chaiteerakij R, Moser CD, Shaleh H, Boakye J, Chen G, Ndzengue A, Li Y, Zhou Y, Huang S, Sinicrope FA, Zou X, Thomas MB, Smith CD, Roberts LR. Antitumor effect of the novel sphingosine kinase 2 inhibitor ABC294640 is enhanced by inhibition of autophagy and by sorafenib in human cholangiocarcinoma cells. Oncotarget. 2016;7:20080-20092. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 572. | Lomas DA, Mahadeva R. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest. 2002;110:1585-1590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 89] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 573. | Huber R, Carrell RW. Implications of the three-dimensional structure of alpha 1-antitrypsin for structure and function of serpins. Biochemistry. 1989;28:8951-8966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 711] [Cited by in F6Publishing: 687] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 574. | Greene CM, Marciniak SJ, Teckman J, Ferrarotti I, Brantly ML, Lomas DA, Stoller JK, McElvaney NG. α1-Antitrypsin deficiency. Nat Rev Dis Primers. 2016;2:16051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 195] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 575. | Lomas DA, Hurst JR, Gooptu B. Update on alpha-1 antitrypsin deficiency: New therapies. J Hepatol. 2016;65:413-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 576. | Rudnick DA, Perlmutter DH. Alpha-1-antitrypsin deficiency: a new paradigm for hepatocellular carcinoma in genetic liver disease. Hepatology. 2005;42:514-521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 107] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 577. | Hazari YM, Bashir A, Habib M, Bashir S, Habib H, Qasim MA, Shah NN, Haq E, Teckman J, Fazili KM. Alpha-1-antitrypsin deficiency: Genetic variations, clinical manifestations and therapeutic interventions. Mutat Res. 2017;773:14-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 578. | Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279:G961-G974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 196] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 579. | Teckman JH, An JK, Loethen S, Perlmutter DH. Fasting in alpha1-antitrypsin deficient liver: constitutive [correction of consultative] activation of autophagy. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1156-G1165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 580. | Marciniak SJ, Lomas DA. Alpha1-antitrypsin deficiency and autophagy. N Engl J Med. 2010;363:1863-1864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 581. | Teckman JH, Mangalat N. Alpha-1 antitrypsin and liver disease: mechanisms of injury and novel interventions. Expert Rev Gastroenterol Hepatol. 2015;9:261-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 582. | Perlmutter DH. Liver injury in alpha1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Invest. 2002;110:1579-1583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 60] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 583. | Teckman JH, An JK, Blomenkamp K, Schmidt B, Perlmutter D. Mitochondrial autophagy and injury in the liver in alpha 1-antitrypsin deficiency. Am J Physiol Gastrointest Liver Physiol. 2004;286:G851-G862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 137] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 584. | Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467-4476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 209] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 585. | Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 146] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 586. | Kaushal S, Annamali M, Blomenkamp K, Rudnick D, Halloran D, Brunt EM, Teckman JH. Rapamycin reduces intrahepatic alpha-1-antitrypsin mutant Z protein polymers and liver injury in a mouse model. Exp Biol Med (Maywood). 2010;235:700-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 587. | Perlmutter DH. The role of autophagy in alpha-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy. 2006;2:258-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 588. | Perlmutter DH. Autophagic disposal of the aggregation-prone protein that causes liver inflammation and carcinogenesis in alpha-1-antitrypsin deficiency. Cell Death Differ. 2009;16:39-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 589. | Lindblad D, Blomenkamp K, Teckman J. Alpha-1-antitrypsin mutant Z protein content in individual hepatocytes correlates with cell death in a mouse model. Hepatology. 2007;46:1228-1235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 590. | Yamamura T, Ohsaki Y, Suzuki M, Shinohara Y, Tatematsu T, Cheng J, Okada M, Ohmiya N, Hirooka Y, Goto H, Fujimoto T. Inhibition of Niemann-Pick-type C1-like1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant α1-antitrypsin Z deposition. Hepatology. 2014;59:1591-1599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 591. | Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, Michalopoulos G, Perlmutter DH. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 453] [Cited by in F6Publishing: 452] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 592. | Pastore N, Blomenkamp K, Annunziata F, Piccolo P, Mithbaokar P, Maria Sepe R, Vetrini F, Palmer D, Ng P, Polishchuk E, Iacobacci S, Polishchuk R, Teckman J, Ballabio A, Brunetti-Pierri N. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med. 2013;5:397-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 593. | Wang Y, Cobanoglu MC, Li J, Hidvegi T, Hale P, Ewing M, Chu AS, Gong Z, Muzumdar R, Pak SC, Silverman GA, Bahar I, Perlmutter DH. An analog of glibenclamide selectively enhances autophagic degradation of misfolded α1-antitrypsin Z. PLoS One. 2019;14:e0209748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 594. | Tang Y, Fickert P, Trauner M, Marcus N, Blomenkamp K, Teckman J. Autophagy induced by exogenous bile acids is therapeutic in a model of α-1-AT deficiency liver disease. Am J Physiol Gastrointest Liver Physiol. 2016;311:G156-G165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 595. | Tang Y, Blomenkamp KS, Fickert P, Trauner M, Teckman JH. NorUDCA promotes degradation of α1-antitrypsin mutant Z protein by inducing autophagy through AMPK/ULK1 pathway. PLoS One. 2018;13:e0200897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 596. | Puls F, Goldschmidt I, Bantel H, Agne C, Bröcker V, Dämmrich M, Lehmann U, Berrang J, Pfister ED, Kreipe HH, Baumann U. Autophagy-enhancing drug carbamazepine diminishes hepatocellular death in fibrinogen storage disease. J Hepatol. 2013;59:626-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 597. | Członkowska A, Litwin T, Dusek P, Ferenci P, Lutsenko S, Medici V, Rybakowski JK, Weiss KH, Schilsky ML. Wilson disease. Nat Rev Dis Primers. 2018;4:21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 386] [Cited by in F6Publishing: 417] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 598. | Polishchuk EV, Merolla A, Lichtmannegger J, Romano A, Indrieri A, Ilyechova EY, Concilli M, De Cegli R, Crispino R, Mariniello M, Petruzzelli R, Ranucci G, Iorio R, Pietrocola F, Einer C, Borchard S, Zibert A, Schmidt HH, Di Schiavi E, Puchkova LV, Franco B, Kroemer G, Zischka H, Polishchuk RS. Activation of Autophagy, Observed in Liver Tissues From Patients With Wilson Disease and From ATP7B-Deficient Animals, Protects Hepatocytes From Copper-Induced Apoptosis. Gastroenterology 2019; 156: 1173-1189. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 599. | Farah BL, Sinha RA, Wu Y, Singh BK, Lim A, Hirayama M, Landau DJ, Bay BH, Koeberl DD, Yen PM. Hepatic mitochondrial dysfunction is a feature of Glycogen Storage Disease Type Ia (GSDIa). Sci Rep. 2017;7:44408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 600. | Farah BL, Landau DJ, Sinha RA, Brooks ED, Wu Y, Fung SYS, Tanaka T, Hirayama M, Bay BH, Koeberl DD, Yen PM. Induction of autophagy improves hepatic lipid metabolism in glucose-6-phosphatase deficiency. J Hepatol. 2016;64:370-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 601. | Cho JH, Kim GY, Pan CJ, Anduaga J, Choi EJ, Mansfield BC, Chou JY. Downregulation of SIRT1 signaling underlies hepatic autophagy impairment in glycogen storage disease type Ia. PLoS Genet. 2017;13:e1006819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 602. | Aki T, Unuma K, Uemura K. Emerging roles of mitochondria and autophagy in liver injury during sepsis. Cell Stress. 2017;1:79-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 603. | Qiu P, Liu Y, Zhang J. Review: the Role and Mechanisms of Macrophage Autophagy in Sepsis. Inflammation. 2019;42:6-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 604. | Chang FY, Chen HC, Chen PJ, Ho MS, Hsieh SL, Lin JC, Liu FT, Sytwu HK. Immunologic aspects of characteristics, diagnosis, and treatment of coronavirus disease 2019 (COVID-19). J Biomed Sci. 2020;27:72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 605. | Lee JP, Foote A, Fan H, Peral de Castro C, Lang T, Jones SA, Gavrilescu N, Mills KH, Leech M, Morand EF, Harris J. Loss of autophagy enhances MIF/macrophage migration inhibitory factor release by macrophages. Autophagy. 2016;12:907-916. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 606. | Thiessen SE, Derese I, Derde S, Dufour T, Pauwels L, Bekhuis Y, Pintelon I, Martinet W, Van den Berghe G, Vanhorebeek I. The Role of Autophagy in Critical Illness-induced Liver Damage. Sci Rep. 2017;7:14150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 607. | Kemp MG. Crosstalk Between Apoptosis and Autophagy: Environmental Genotoxins, Infection, and Innate Immunity. J Cell Death. 2017;9:1179670716685085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 608. | Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, Reinert L, Cai Y, Jensen SB, Skouboe MK, Nyengaard JR, Thompson CB, Lebbink RJ, Sen GC, van Loo G, Nielsen R, Komatsu M, Nejsum LN, Jakobsen MR, Gyrd-Hansen M, Paludan SR. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 284] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 609. | Hu Q, Knight PH, Ren Y, Ren H, Zheng J, Wu X, Ren J, Sawyer RG. The emerging role of stimulator of interferons genes signaling in sepsis: Inflammation, autophagy, and cell death. Acta Physiol (Oxf). 2019;225:e13194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 610. | Lalazar G, Ilyas G, Malik SA, Liu K, Zhao E, Amir M, Lin Y, Tanaka KE, Czaja MJ. Autophagy confers resistance to lipopolysaccharide-induced mouse hepatocyte injury. Am J Physiol Gastrointest Liver Physiol. 2016;311:G377-G386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 611. | Oami T, Watanabe E, Hatano M, Teratake Y, Fujimura L, Sakamoto A, Ito C, Toshimori K, Swanson PE, Oda S. Blocking Liver Autophagy Accelerates Apoptosis and Mitochondrial Injury in Hepatocytes and Reduces Time to Mortality in a Murine Sepsis Model. Shock. 2018;50:427-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 612. | Xiong X, Ren Y, Cui Y, Li R, Wang C, Zhang Y. Obeticholic acid protects mice against lipopolysaccharide-induced liver injury and inflammation. Biomed Pharmacother. 2017;96:1292-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 613. | Xing W, Yang L, Peng Y, Wang Q, Gao M, Yang M, Xiao X. Ginsenoside Rg3 attenuates sepsis-induced injury and mitochondrial dysfunction in liver via AMPK-mediated autophagy flux. Biosci Rep. 2017;37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 614. | Inata Y, Kikuchi S, Samraj RS, Hake PW, O'Connor M, Ledford JR, O'Connor J, Lahni P, Wolfe V, Piraino G, Zingarelli B. Autophagy and mitochondrial biogenesis impairment contribute to age-dependent liver injury in experimental sepsis: dysregulation of AMP-activated protein kinase pathway. FASEB J. 2018;32:728-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 615. | Ying L, Zhao GJ, Wu Y, Ke HL, Hong GL, Zhang H, Dong N, Wu Y, Yao YM, Lu ZQ. Mitofusin 2 Promotes Apoptosis of CD4+ T Cells by Inhibiting Autophagy in Sepsis. Mediators Inflamm. 2017;2017:4926205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 616. | Neumann Y, Bruns SA, Rohde M, Prajsnar TK, Foster SJ, Schmitz I. Intracellular Staphylococcus aureus eludes selective autophagy by activating a host cell kinase. Autophagy. 2016;12:2069-2084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 617. | Yin X, Xin H, Mao S, Wu G, Guo L. The Role of Autophagy in Sepsis: Protection and Injury to Organs. Front Physiol. 2019;10:1071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 618. | Sydor S, Manka P, Best J, Jafoui S, Sowa JP, Zoubek ME, Hernandez-Gea V, Cubero FJ, Kälsch J, Vetter D, Fiel MI, Hoshida Y, Bian CB, Nelson LJ, Moshage H, Faber KN, Paul A, Baba HA, Gerken G, Friedman SL, Canbay A, Bechmann LP. Krüppel-like factor 6 is a transcriptional activator of autophagy in acute liver injury. Sci Rep. 2017;7:8119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 619. | Baulies A, Ribas V, Núñez S, Torres S, Alarcón-Vila C, Martínez L, Suda J, Ybanez MD, Kaplowitz N, García-Ruiz C, Fernández-Checa JC. Lysosomal Cholesterol Accumulation Sensitizes To Acetaminophen Hepatotoxicity by Impairing Mitophagy. Sci Rep. 2015;5:18017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 620. | James LP, Alonso EM, Hynan LS, Hinson JA, Davern TJ, Lee WM, Squires RH; Pediatric Acute Liver Failure Study Group. Detection of acetaminophen protein adducts in children with acute liver failure of indeterminate cause. Pediatrics. 2006;118:e676-e681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 65] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 621. | Xie Y, McGill MR, Du K, Dorko K, Kumer SC, Schmitt TM, Ding WX, Jaeschke H. Mitochondrial protein adducts formation and mitochondrial dysfunction during N-acetyl-m-aminophenol (AMAP)-induced hepatotoxicity in primary human hepatocytes. Toxicol Appl Pharmacol. 2015;289:213-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 622. | Qiu Y, Benet LZ, Burlingame AL. Identification of the hepatic protein targets of reactive metabolites of acetaminophen in vivo in mice using two-dimensional gel electrophoresis and mass spectrometry. J Biol Chem. 1998;273:17940-17953. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 224] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 623. | McGill MR, Lebofsky M, Norris HR, Slawson MH, Bajt ML, Xie Y, Williams CD, Wilkins DG, Rollins DE, Jaeschke H. Plasma and liver acetaminophen-protein adduct levels in mice after acetaminophen treatment: dose-response, mechanisms, and clinical implications. Toxicol Appl Pharmacol. 2013;269:240-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 180] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 624. | Heard KJ, Green JL, James LP, Judge BS, Zolot L, Rhyee S, Dart RC. Acetaminophen-cysteine adducts during therapeutic dosing and following overdose. BMC Gastroenterol. 2011;11:20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 625. | Ni HM, McGill MR, Chao X, Du K, Williams JA, Xie Y, Jaeschke H, Ding WX. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J Hepatol. 2016;65:354-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 147] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 626. | Lin Z, Wu F, Lin S, Pan X, Jin L, Lu T, Shi L, Wang Y, Xu A, Li X. Adiponectin protects against acetaminophen-induced mitochondrial dysfunction and acute liver injury by promoting autophagy in mice. J Hepatol. 2014;61:825-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 627. | Igusa Y, Yamashina S, Izumi K, Inami Y, Fukada H, Komatsu M, Tanaka K, Ikejima K, Watanabe S. Loss of autophagy promotes murine acetaminophen hepatotoxicity. J Gastroenterol. 2012;47:433-443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 628. | Sun Y, Li TY, Song L, Zhang C, Li J, Lin ZZ, Lin SC, Lin SY. Liver-specific deficiency of unc-51 Like kinase 1 and 2 protects mice from acetaminophen-induced liver injury. Hepatology. 2018;67:2397-2413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 629. | Shan S, Shen Z, Zhang C, Kou R, Xie K, Song F. Mitophagy protects against acetaminophen-induced acute liver injury in mice through inhibiting NLRP3 inflammasome activation. Biochem Pharmacol. 2019;169:113643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 630. | Hua D, Ju Z, Gan X, Wang Q, Luo C, Gu J, Yu Y. Human amniotic mesenchymal stromal cells alleviate acute liver injury by inhibiting the pro-inflammatory response of liver resident macrophage through autophagy. Ann Transl Med. 2019;7:392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 631. | Wu Z, Han M, Chen T, Yan W, Ning Q. Acute liver failure: mechanisms of immune-mediated liver injury. Liver Int. 2010;30:782-794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 202] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 632. | Dechêne A, Sowa JP, Gieseler RK, Jochum C, Bechmann LP, El Fouly A, Schlattjan M, Saner F, Baba HA, Paul A, Dries V, Odenthal M, Gerken G, Friedman SL, Canbay A. Acute liver failure is associated with elevated liver stiffness and hepatic stellate cell activation. Hepatology. 2010;52:1008-1016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 633. | Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123:1902-1910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 527] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 634. | Shen K, Chang W, Gao X, Wang H, Niu W, Song L, Qin X. Depletion of activated hepatic stellate cell correlates with severe liver damage and abnormal liver regeneration in acetaminophen-induced liver injury. Acta Biochim Biophys Sin (Shanghai). 2011;43:307-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 635. | He Y, Jin L, Wang J, Yan Z, Chen T, Zhao Y. Mechanisms of fibrosis in acute liver failure. Liver Int. 2015;35:1877-1885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 636. | Langer DA, Das A, Semela D, Kang-Decker N, Hendrickson H, Bronk SF, Katusic ZS, Gores GJ, Shah VH. Nitric oxide promotes caspase-independent hepatic stellate cell apoptosis through the generation of reactive oxygen species. Hepatology. 2008;47:1983-1993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 637. | Jin L, Gao H, Wang J, Yang S, Wang J, Liu J, Yang Y, Yan T, Chen T, Zhao Y, He Y. Role and regulation of autophagy and apoptosis by nitric oxide in hepatic stellate cells during acute liver failure. Liver Int. 2017;37:1651-1659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 638. | Amir M, Zhao E, Fontana L, Rosenberg H, Tanaka K, Gao G, Czaja MJ. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 2013;20:878-887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 639. | Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, Karin M. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896-910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 835] [Cited by in F6Publishing: 818] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 640. | Han J, Bae J, Choi CY, Choi SP, Kang HS, Jo EK, Park J, Lee YS, Moon HS, Park CG, Lee MS, Chun T. Autophagy induced by AXL receptor tyrosine kinase alleviates acute liver injury via inhibition of NLRP3 inflammasome activation in mice. Autophagy. 2016;12:2326-2343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 641. | Wang Q, Wei S, Zhou S, Qiu J, Shi C, Liu R, Zhou H, Lu L. Hyperglycemia aggravates acute liver injury by promoting liver-resident macrophage NLRP3 inflammasome activation via the inhibition of AMPK/mTOR-mediated autophagy induction. Immunol Cell Biol. 2020;98:54-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 642. | Zhou S, Gu J, Liu R, Wei S, Wang Q, Shen H, Dai Y, Zhou H, Zhang F, Lu L. Spermine Alleviates Acute Liver Injury by Inhibiting Liver-Resident Macrophage Pro-Inflammatory Response Through ATG5-Dependent Autophagy. Front Immunol. 2018;9:948. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 643. | Kim JS, Wang JH, Biel TG, Kim DS, Flores-Toro JA, Vijayvargiya R, Zendejas I, Behrns KE. Carbamazepine suppresses calpain-mediated autophagy impairment after ischemia/reperfusion in mouse livers. Toxicol Appl Pharmacol. 2013;273:600-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 644. | Biel TG, Lee S, Flores-Toro JA, Dean JW, Go KL, Lee MH, Law BK, Law ME, Dunn WA Jr, Zendejas I, Behrns KE, Kim JS. Sirtuin 1 suppresses mitochondrial dysfunction of ischemic mouse livers in a mitofusin 2-dependent manner. Cell Death Differ. 2016;23:279-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 645. | Evankovich J, Zhang R, Cardinal JS, Zhang L, Chen J, Huang H, Beer-Stolz D, Billiar TR, Rosengart MR, Tsung A. Calcium/calmodulin-dependent protein kinase IV limits organ damage in hepatic ischemia-reperfusion injury through induction of autophagy. Am J Physiol Gastrointest Liver Physiol. 2012;303:G189-G198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 646. | Kim JS, Nitta T, Mohuczy D, O'Malley KA, Moldawer LL, Dunn WA Jr, Behrns KE. Impaired autophagy: A mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725-1736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 647. | Fang H, Liu A, Dahmen U, Dirsch O. Dual role of chloroquine in liver ischemia reperfusion injury: reduction of liver damage in early phase, but aggravation in late phase. Cell Death Dis. 2013;4:e694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 648. | Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 232] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 649. | Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011;108:11121-11126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 276] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 650. | Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T, Schutkowski M, Pellegrini L, Sansone L, Villanova L, Runci A, Pucci B, Morgante E, Fini M, Mai A, Russo MA, Tafani M. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11:253-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 213] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 651. | Soria LR, Allegri G, Melck D, Pastore N, Annunziata P, Paris D, Polishchuk E, Nusco E, Thöny B, Motta A, Häberle J, Ballabio A, Brunetti-Pierri N. Enhancement of hepatic autophagy increases ureagenesis and protects against hyperammonemia. Proc Natl Acad Sci USA. 2018;115:391-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 652. | Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 237] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 653. | Golbabapour S, Bagheri-Lankarani K, Ghavami S, Geramizadeh B. Autoimmune Hepatitis and Stellate Cells: An Insight into the Role of Autophagy. Curr Med Chem. 2020;27:6073-6095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 654. | Feng Q, Yao J, Zhou G, Xia W, Lyu J, Li X, Zhao T, Zhang G, Zhao N, Yang J. Quantitative Proteomic Analysis Reveals That Arctigenin Alleviates Concanavalin A-Induced Hepatitis Through Suppressing Immune System and Regulating Autophagy. Front Immunol. 2018;9:1881. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 655. | Fan X, Men R, Wang H, Shen M, Wang T, Ye T, Luo X, Yang L. Methylprednisolone Decreases Mitochondria-Mediated Apoptosis and Autophagy Dysfunction in Hepatocytes of Experimental Autoimmune Hepatitis Model via the Akt/mTOR Signaling. Front Pharmacol. 2019;10:1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 656. | Fan X, Men R, Huang C, Shen M, Wang T, Ghnewa Y, Ma Y, Ye T, Yang L. Critical roles of conventional dendritic cells in autoimmune hepatitis via autophagy regulation. Cell Death Dis. 2020;11:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 657. | Szekerczés T, Gógl A, Illyés I, Mandl J, Borka K, Kiss A, Schaff Z, Lendvai G, Werling K. Autophagy, Mitophagy and MicroRNA Expression in Chronic Hepatitis C and Autoimmune Hepatitis. Pathol Oncol Res. 2020;26:2143-2151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 658. | Kwak BJ, Choi HJ, Kim OH, Kim KH, You YK, Lee TY, Ahn J, Kim SJ. The Role of Phospho-c-Jun N-Terminal Kinase Expression on hepatocyte Necrosis and Autophagy in the Cholestatic Liver. J Surg Res. 2019;241:254-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 659. | Huang YH, Yang YL, Huang FC, Tiao MM, Lin YC, Tsai MH, Wang FS. MicroRNA-29a mitigation of endoplasmic reticulum and autophagy aberrance counteracts in obstructive jaundice-induced fibrosis in mice. Exp Biol Med (Maywood). 2018;243:13-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 660. | Kim S, Han SY, Yu KS, Han D, Ahn HJ, Jo JE, Kim JH, Shin J, Park HW. Impaired autophagy promotes bile acid-induced hepatic injury and accumulation of ubiquitinated proteins. Biochem Biophys Res Commun. 2018;495:1541-1547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 661. | Khambu B, Li T, Yan S, Yu C, Chen X, Goheen M, Li Y, Lin J, Cummings OW, Lee YA, Friedman S, Dong Z, Feng GS, Wu S, Yin XM. Hepatic Autophagy Deficiency Compromises Farnesoid X Receptor Functionality and Causes Cholestatic Injury. Hepatology. 2019;69:2196-2213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 662. | Gao L, Lv G, Guo X, Jing Y, Han Z, Zhang S, Sun K, Li R, Yang Y, Wei L. Activation of autophagy protects against cholestasis-induced hepatic injury. Cell Biosci. 2014;4:47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 663. | Woolbright BL, Jaeschke H. Novel insight into mechanisms of cholestatic liver injury. World J Gastroenterol. 2012;18:4985-4993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 148] [Cited by in F6Publishing: 143] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 664. | Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. Modulation of the microenvironment by senescent biliary epithelial cells may be involved in the pathogenesis of primary biliary cirrhosis. J Hepatol. 2010;53:318-325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 665. | Sasaki M, Nakanuma Y. Novel approach to bile duct damage in primary biliary cirrhosis: participation of cellular senescence and autophagy. Int J Hepatol. 2012;2012:452143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 666. | Manley S, Ni HM, Kong B, Apte U, Guo G, Ding WX. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci. 2014;137:478-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 667. | Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. Increased expression of mitochondrial proteins associated with autophagy in biliary epithelial lesions in primary biliary cirrhosis. Liver Int. 2013;33:312-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 668. | Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. Autophagy mediates the process of cellular senescence characterizing bile duct damages in primary biliary cirrhosis. Lab Invest. 2010;90:835-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 669. | van de Graaf S, Beuers U. Autophagy - another piece of the puzzle towards understanding primary biliary cirrhosis? Liver Int. 2014;34:481-483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 670. | Nakanuma Y, Sasaki M, Harada K. Autophagy and senescence in fibrosing cholangiopathies. J Hepatol. 2015;62:934-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 671. | Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. Autophagy may precede cellular senescence of bile ductular cells in ductular reaction in primary biliary cirrhosis. Dig Dis Sci. 2012;57:660-666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 672. | Sasaki M, Nakanuma Y. Biliary epithelial apoptosis, autophagy, and senescence in primary biliary cirrhosis. Hepat Res Treat. 2010;2010:205128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 673. | Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavaré S, Arakawa S, Shimizu S, Watt FM, Narita M. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798-803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 741] [Cited by in F6Publishing: 795] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 674. | Sasaki M, Ikeda H, Haga H, Manabe T, Nakanuma Y. Frequent cellular senescence in small bile ducts in primary biliary cirrhosis: a possible role in bile duct loss. J Pathol. 2005;205:451-459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 92] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 675. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: The diagnosis and management of patients with primary biliary cholangitis. J Hepatol. 2017;67:145-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 641] [Cited by in F6Publishing: 766] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 676. | Lindor KD, Bowlus CL, Boyer J, Levy C, Mayo M. Primary Biliary Cholangitis: 2018 Practice Guidance from the American Association for the Study of Liver Diseases. Hepatology. 2019;69:394-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 317] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 677. | Gossard AA, Lindor KD. Current and promising therapy for primary biliary cholangitis. Expert Opin Pharmacother. 2019;20:1161-1167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 678. | Sasaki M, Nakanuma Y. Bile Acids and Deregulated Cholangiocyte Autophagy in Primary Biliary Cholangitis. Dig Dis. 2017;35:210-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 679. | Mueller M, Thorell A, Claudel T, Jha P, Koefeler H, Lackner C, Hoesel B, Fauler G, Stojakovic T, Einarsson C, Marschall HU, Trauner M. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol. 2015;62:1398-1404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 680. | Panzitt K, Jungwirth E, Krones E, Lee JM, Pollheimer M, Thallinger GG, Kolb-Lenz D, Xiao R, Thorell A, Trauner M, Fickert P, Marschall HU, Moore DD, Wagner M. FXR-dependent Rubicon induction impairs autophagy in models of human cholestasis. J Hepatol. 2020;72:1122-1131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 681. | Wagner M, Zollner G, Trauner M. Nuclear receptors in liver disease. Hepatology. 2011;53:1023-1034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 211] [Article Influence: 16.2] [Reference Citation Analysis (0)] |