Copyright
©The Author(s) 2020.
World J Hepatol. Dec 27, 2020; 12(12): 1211-1227
Published online Dec 27, 2020. doi: 10.4254/wjh.v12.i12.1211
Published online Dec 27, 2020. doi: 10.4254/wjh.v12.i12.1211
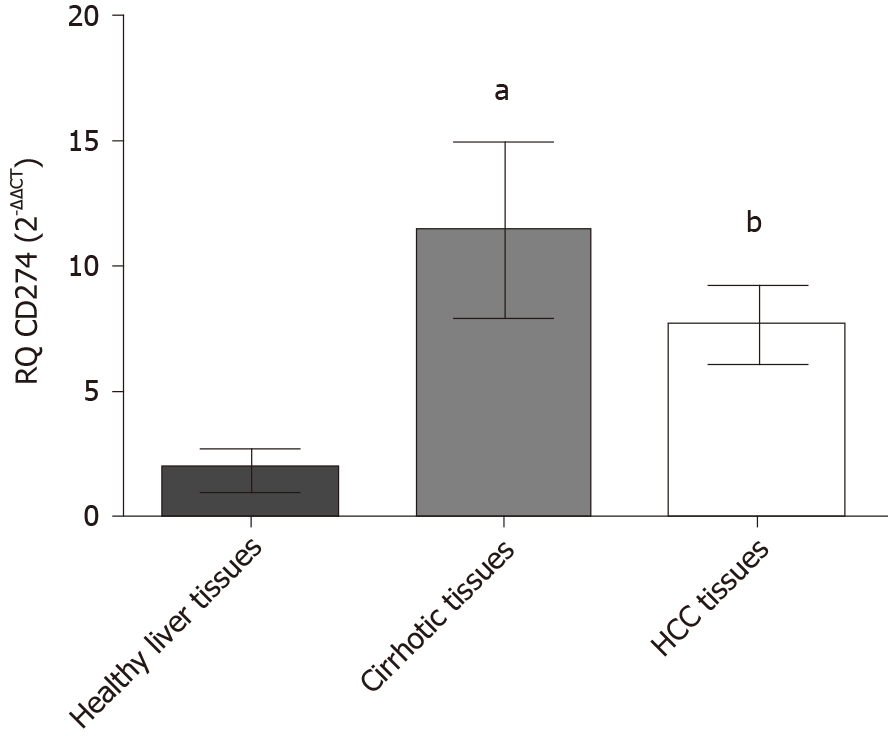
Figure 1 Relative expression level of programmed death ligand 1 in liver tissues.
Endogenous programmed death ligand 1 expression profile was analyzed in hepatocellular carcinoma patients, cirrhotic and healthy controls using quantified real-time polymerase chain reaction and normalized to B2M as an internal control (housekeeping gene). Screening of programmed death ligand 1 showed that it was enhanced in cirrhotic biopsies (aP < 0.05) and hepatocellular carcinoma biopsies (bP < 0.01) compared to healthy controls. HCC: Hepatocellular carcinoma.
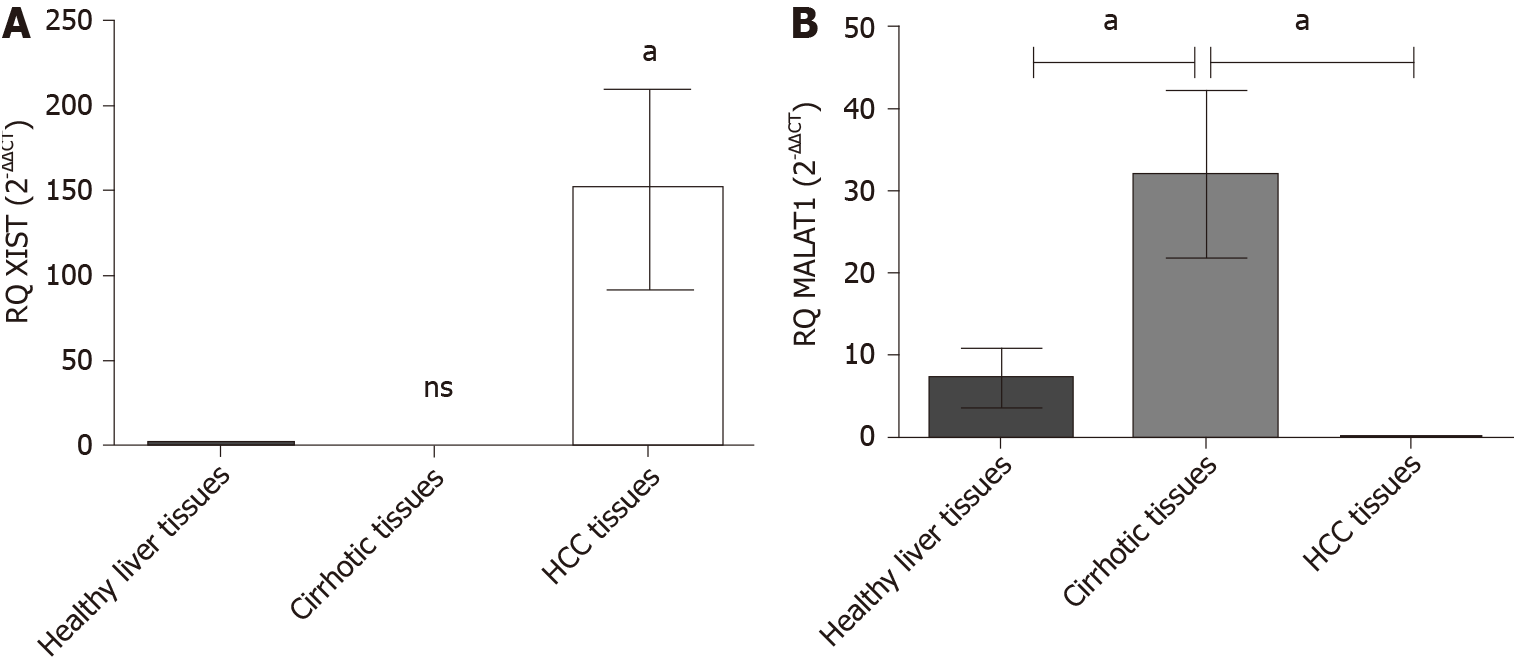
Figure 2 Expression profile of lnc-ribonucleic acid X-inactive specific transcript and MALAT-1 in hepatocellular carcinoma tissues.
Endogenous X-inactive specific transcript and MALAT-1 lnc-ribonucleic acids expression profile was analyzed in hepatocellular carcinoma (HCC) patients and healthy controls using quantified real-time polymerase chain reaction and normalized to B2M as an endogenous control. A: X-inactive specific transcript lnc-ribonucleic acid showed a significant upregulation in HCC biopsies (P = 0.048); and B: MALAT-1 was significantly down regulated in HCC biopsies (P = 0.043); however, it showed elevated expression in cirrhotic biopsies (P = 0.0136). aP < 0.05. HCC: Hepatocellular carcinoma.
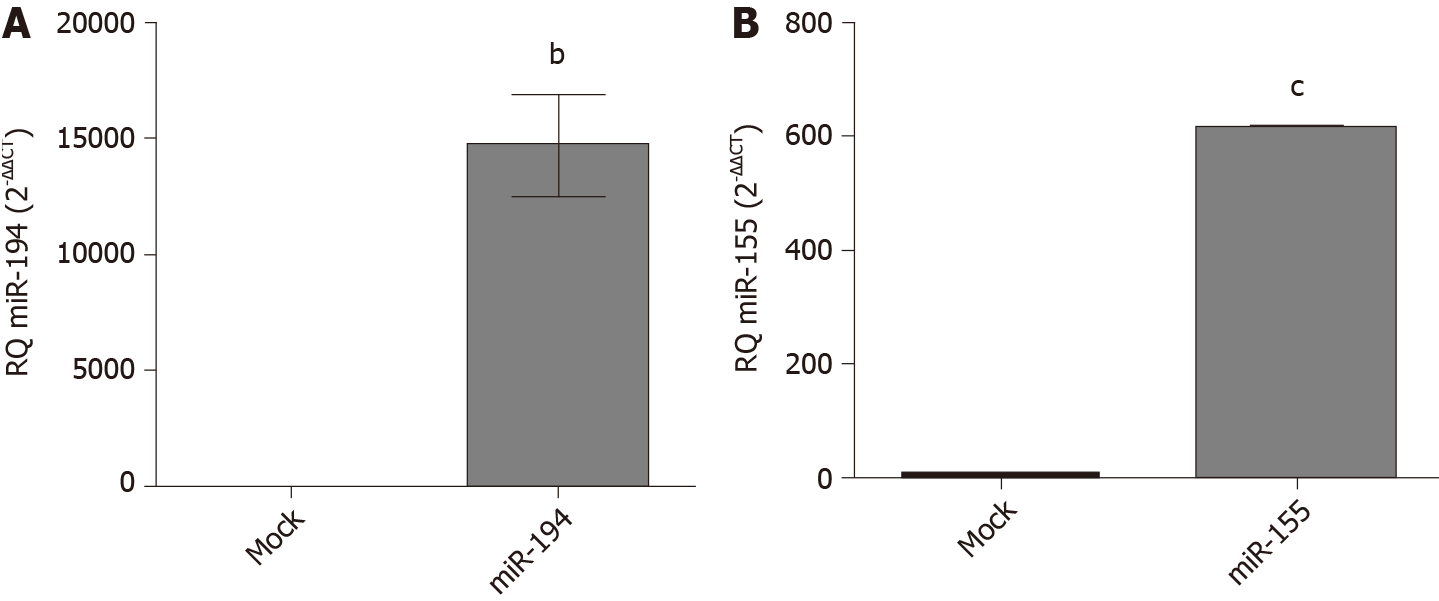
Figure 3 Transfection efficiency in Huh-7 cells with miR194-5p and miR-155-5p oligonucleotides.
Using quantified real-time-polymerase chain reaction, the transfection efficiency was determined in both mimicked and mock cells 48 h post-transfection for each of the respective mi-ribonucleic acids. MiR-194-5p and miR-155-5p were normalized to RNU6B as an endogenous control. A: Mimicking of miR-194-5p; and B: miR-155-5p resulted in an increase in each of the respective miRNAs, (P = 0.0026) and (P < 0.0001), respectively. The expression levels were compared with the unpaired Student’s t-test. bP < 0.01 and cP < 0.001.
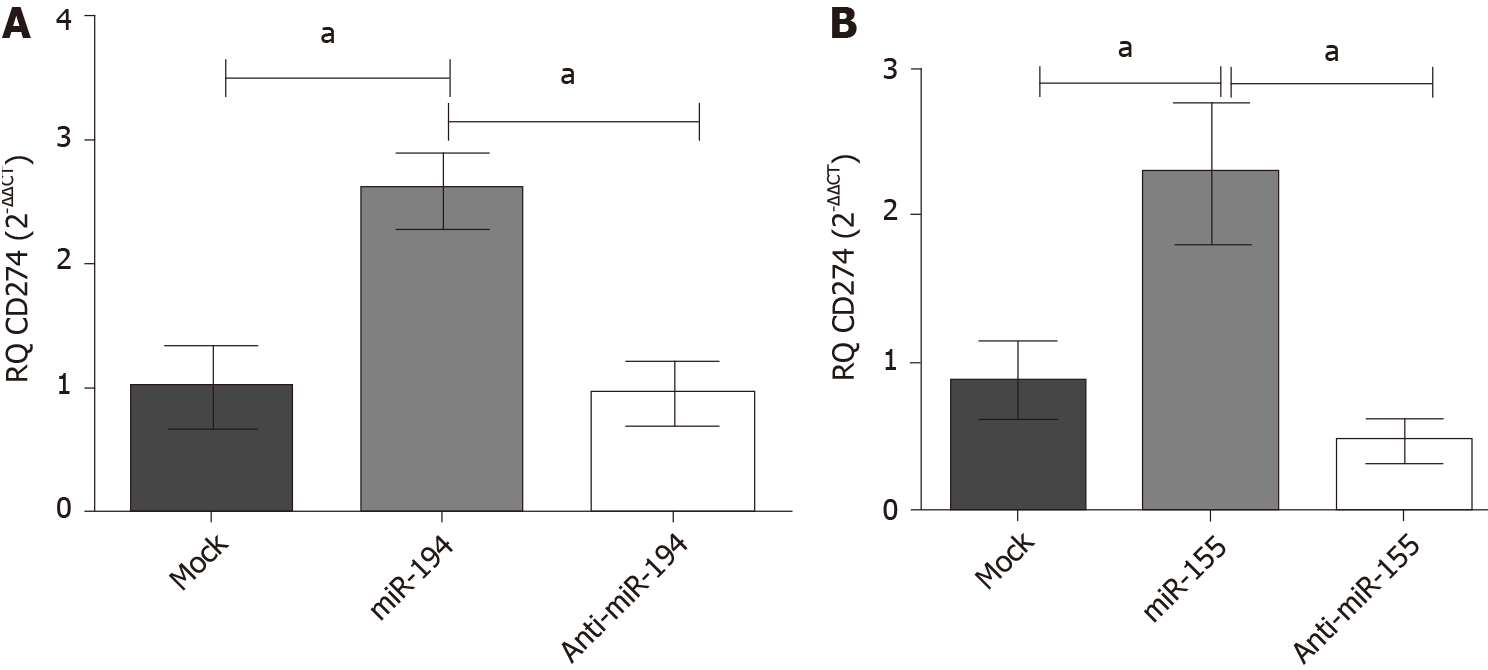
Figure 4 Impact of miR-194-5p and miR-155-5p on programmed death ligand 1 transcript expression in Huh-7 cells.
Following ectopic expression manipulation of (A) miR-194-5p and (B) miR-155-5p in Huh-7 cells, programmed death ligand 1 (PD-L1) transcript expression was assessed using quantified real-time polymerase chain reaction and normalized to B2M as an endogenous control. Mimicking of each of the respective miRNAs resulted in significant upregulation of PD-L1 compared to mock untransfected cells (P = 0.0219) and (P = 0.0209), respectively. On the contrary, PD-L1 transcript expression was significantly downregulated by antagonizing each of the miRNAs in comparison with the mock untransfected cells. aP < 0.05.
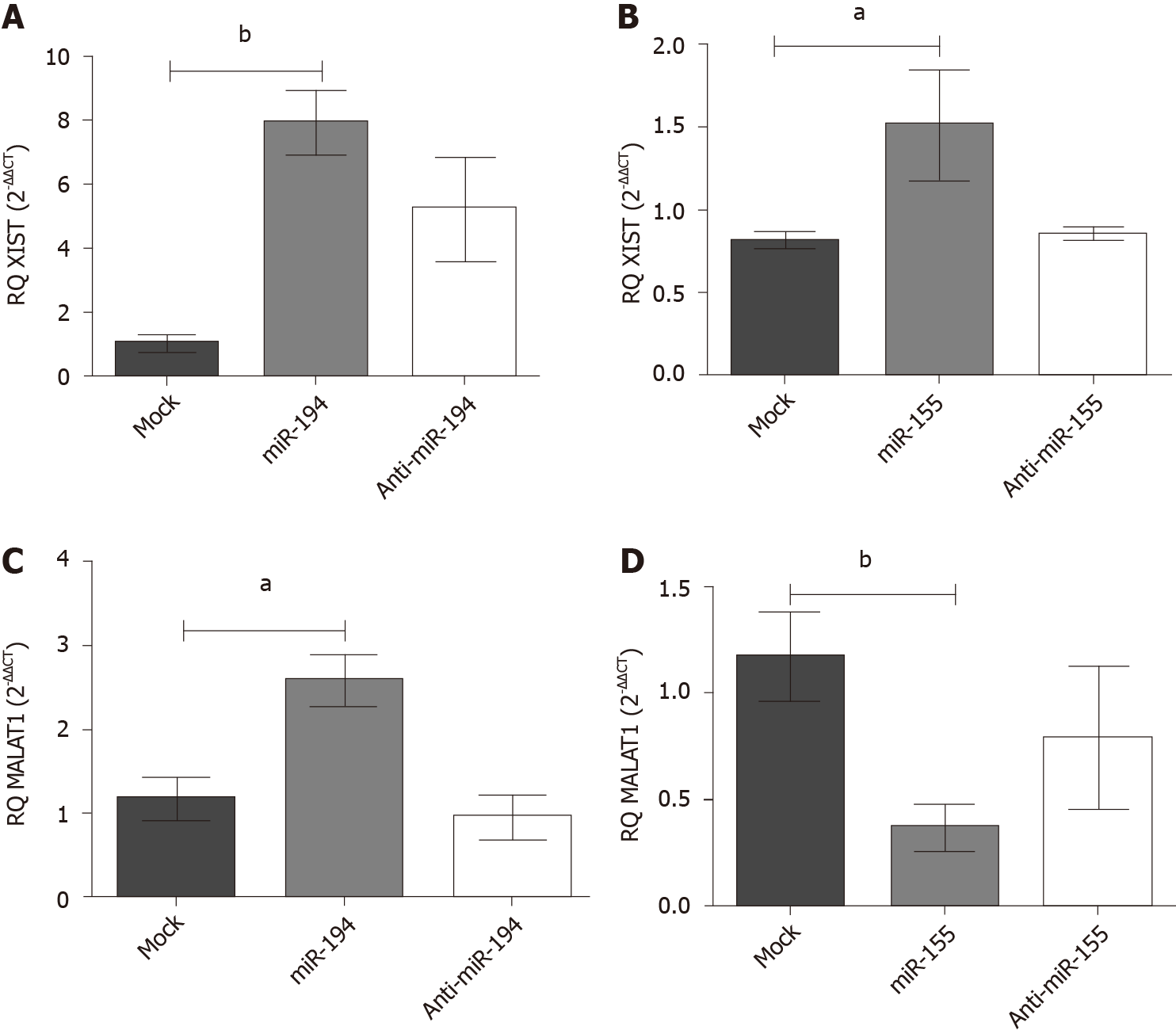
Figure 5 Impact of miR-155-5p and miR-194-5p on lnc-ribonucleic acids X-inactive specific transcript and MALAT-1 expression in Huh-7 cells.
Expression levels of lnc-ribonucleic acids X-inactive specific transcript and MALAT-1 were assessed following transfection of miR-194-5p and miR-155-5p oligomirs using quantified real-time polymerase chain reaction and normalized to the endogenous B2M as a housekeeping gene. A: Ectopic expression of both miR-155 and mir-194 resulted in significant upregulation of X-inactive specific transcript expression, (P = 0.0477) and (P = 0.0026) respectively, compared to mock cells; and B: However, a paradoxical effect of mimicking miR-194-5p and miR-155-5p on MALAT-1 expression profile was observed, as miR-194-5p stimulated the upregulation of MALAT-1 expression (P = 0.0135), whereas mimicking miR-155-5p induced downregulation of MALAT-1 expression in comparison to mock cells (P = 0.0053). aP < 0.05 and bP < 0.01.
Figure 6 Impact of knockdown of long non-coding ribonucleic acids MALAT-1, X-inactive specific transcript and the negative regulator of X-inactive specific transcript on programmed death ligand 1 expression in Huh-7 cells.
Using quantified real-time polymerase chain reaction, the expression profile of programmed death ligand 1 (PD-L1) transcript was determined following transfection of Huh-7 cells with each of the siRNAs for lncRNAs MALAT-1, X-inactive specific transcript and the negative regulator of X-inactive specific transcript. Values were normalized to an endogenous housekeeping gene B2M. PD-L1 expression was induced following knockdown of MALAT-1 (P = 0.0010) compared to mock cells. On the contrary, following down regulation of the negative regulator of X-inactive specific transcript, PD-L1 transcript expression was significantly induced (P = 0.0358) compared to mock cells. PD-L1 expression level was totally unaffected by knocking down MALAT-1 in comparison with mock cells. TSIX: The negative regulator of X-inactive specific transcript.
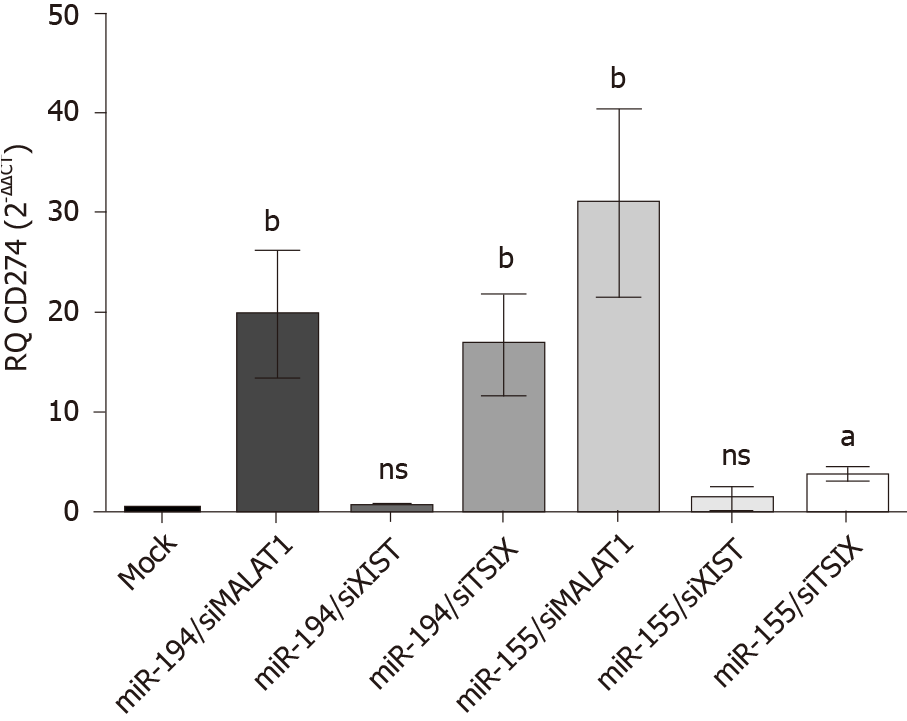
Figure 7 Net impact of ectopic miR-194-5p and miR-155-5p expression on programmed death ligand 1 expression in the presence of siRNA of lnc-ribonucleic acids X-inactive specific transcript, the negative regulator of X-inactive specific transcript and MALAT-1.
The impact of both tumour suppressor miR-194-5p and oncogenic miR-155-5p on programmed death ligand 1 (PD-L1) expression in Huh7 cells in the presence of long non-coding ribonucleic acid X-inactive specific transcript (XIST), the negative regulator of X-inactive specific transcript and MALAT-1 siRNA was similar. Both miRNAs induced upregulation of PD-L1 expression when each was accompanied by MALAT-1 siRNA, while knockdown of the long non-coding ribonucleic acid the negative regulator of X-inactive specific transcript, i.e. upregulation of XIST expression, in the presence of miR-155-5p and miR-194-5p mimics down regulated PD-L1 expression. Ectopic expression of each of the miRNAs with knockdown of XIST had no net impact on PD-L1 expression. aP < 0.05 and bP < 0.01. TSIX: The negative regulator of X-inactive specific transcript.
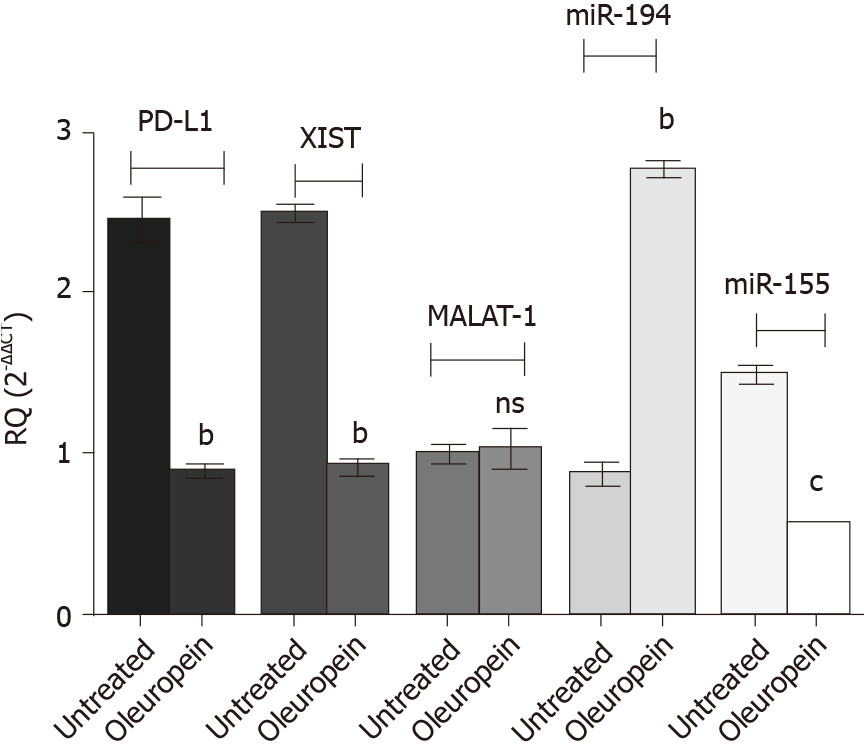
Figure 8 Impact of oleuropein on the expression profile of the study key players.
The expression pattern of programmed death ligand 1 (PD-L1), micro ribonucleic acids 194-5p and 155-5p as well as long non-coding ribonucleic acids X-inactive specific transcript (XIST) and MALAT-1 were assessed following oleuropein treatment. Values for micro ribonucleic acids were normalized to RNU6B while that for PD-L1, XIST and MALAT-1 were normalized to B2M. Enhanced expression of miR-194-5p was observed following oleuropein treatment (P = 0.0022). However, PD-L1, XIST and miR-155-5p expression pattern were down regulated, (P = 0.0011), (P = 0.0020) and (P = 0.0001), respectively. Oleuropein did not show any significant impact on MALAT-1 expression profile. bP < 0.01 and cP < 0.001.
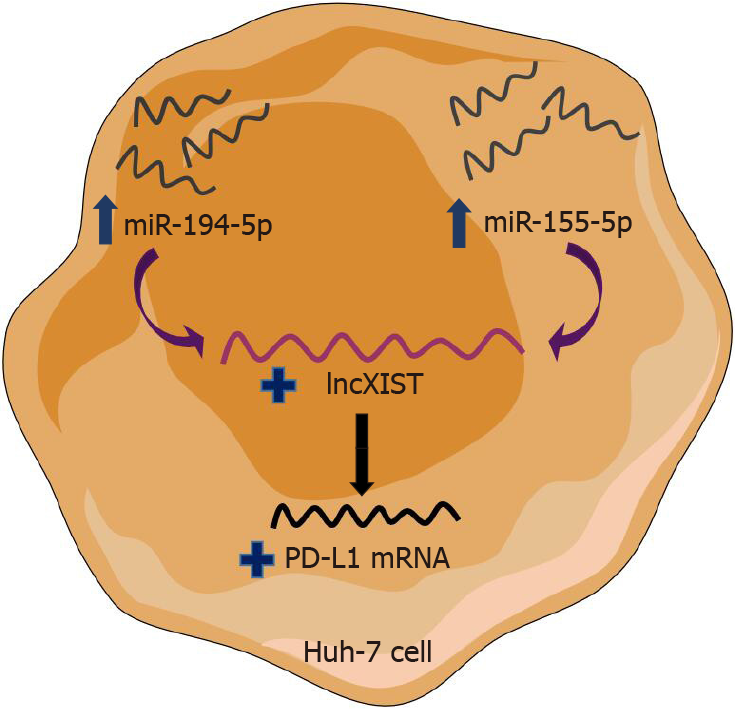
Figure 9 Schematic representation of the shared pathway between miR-155-5p and miR-194-5p.
This article highlights the novel shared upstream regulatory signaling pathways for programmed cell-death protein 1/programmed death ligand 1 immune checkpoint between paradoxically acting miR-194-5p and miR-155-5p, through lnc X-inactive specific transcript expression modulation. Mimicking of tumor suppressor miR-194-5p as well as oncomiR-155-5p in the Huh-7 cell line showed the same upregulation pattern of X-inactive specific transcript. X-inactive specific transcript was proposed then to be an intermediate player whose upregulation derived the increase in programmed death ligand 1 transcript abundance.
- Citation: Atwa SM, Handoussa H, Hosny KM, Odenthal M, El Tayebi HM. Pivotal role of long non-coding ribonucleic acid-X-inactive specific transcript in regulating immune checkpoint programmed death ligand 1 through a shared pathway between miR-194-5p and miR-155-5p in hepatocellular carcinoma. World J Hepatol 2020; 12(12): 1211-1227
- URL: https://www.wjgnet.com/1948-5182/full/v12/i12/1211.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i12.1211