修回日期: 2003-08-20
接受日期: 2003-09-01
在线出版日期: 2003-10-15
研究丙型肝炎病毒核糖体插入位点启动翻译蛋白质功能, 构建双顺反子真核表达载体.
逆转录PCR(RT-PCR)扩增丙型肝炎基因组中核糖体插入位点序列(IRES), 定向克隆到pcDNA3-S质粒多克隆酶切位点中. 在IRES下游克隆乙型肝炎病毒核心基因, 测序鉴定后获得的质粒经脂质体转染hepG2细胞, 间接免疫荧光染色和Western-blot检测.
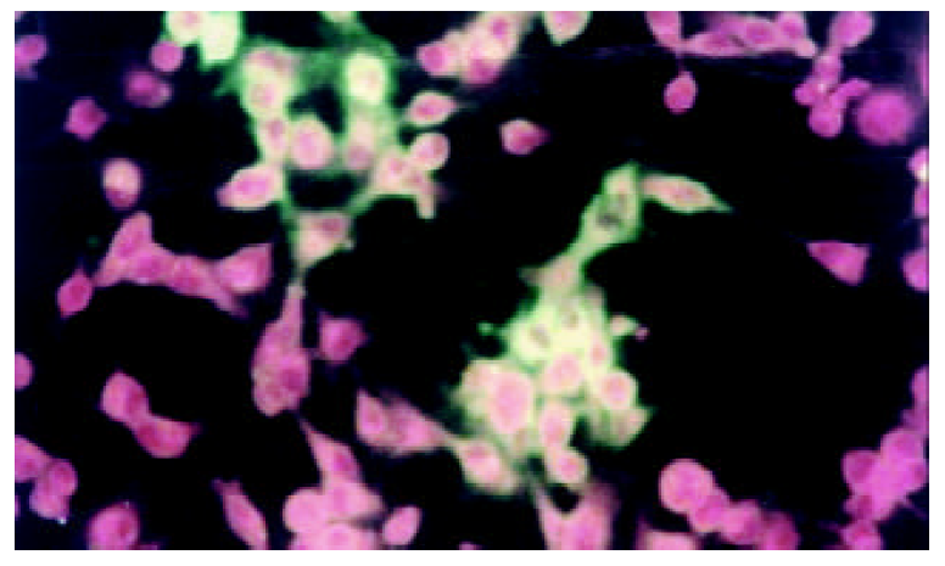
在间接免疫荧光染色后, 荧光显微镜下可见乙型肝炎病毒S基因和核心基因表达. 转染细胞破碎后免疫沉淀检测和图像分析表明, 两种基因有相似的表达量.
人为截断HCV 5'NTR区17个碱基, 并不影响IRES对下游基因蛋白质翻译, 为成功构建了含丙型肝炎病毒 IRES的双顺反子表达载体打下基础.
引文著录: 阚全程, 余祖江, 雷延昌, 杨东亮, 郝连杰. 人工构建含丙型肝炎病毒核糖体插入位点的双顺反子表达载体. 世界华人消化杂志 2003; 11(10): 1520-1523
Revised: August 20, 2003
Accepted: September 1, 2003
Published online: October 15, 2003
To study the function of hepatitis C virus(HCV) internal ribosome entry site (IRES), and to construct biscistronic vector.
After amplifying HCV IRES by reverse-transcription PCR (RT-PCR), the products were cloned into pcDNA3-S upstream hepatitis B virus (HBV) surface gene. HBV core gene was cloned following HCV IRES. After determination by PCR and sequencing, we acquired plasmids containing HBV S, C gene and HCV IRES, which were named as plasmids pcDNA3-SIC. PcDNA3-SIC were transfected into HepG2 cells and detected by immunofluorescence assay and Western blot.
HBV surface gene and core gene were both expressed in hepG2 cells, which were detected by immunofluorescence assay and confirmed by Western blot.
The 17 nt of 5' nontranslated RNA in HCV IRES had no effect on driving downstream gene expression itself and could be used in the biscistronic vector that drove two genes expression.
- Citation: Kan QC, Yu ZJ, Lei YC, Yang DL, Hao LJ. Artificially constructed biscistronic vector containing hepatitis C virus internal ribosome entry site. Shijie Huaren Xiaohua Zazhi 2003; 11(10): 1520-1523
- URL: https://www.wjgnet.com/1009-3079/full/v11/i10/1520.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v11.i10.1520
在引起慢性病毒性肝炎的病原中, 丙型肝炎病毒(hepatitis C virus, HCV)是重要的病原之一[1-7]. 丙型肝炎的慢性化机制虽然和乙型肝炎有许多相似之处, 如免疫耐受的形成, 细胞凋亡等[8-10]. 但也有许多不同之处, 如HCV基因组具有高变性, 在血液中间断性出现等原因, 不易被免疫系统识别而造成患者慢性化[11-14]. 最近有研究表明, HCV核心蛋白可以和其IRES位点相结合, 下调HCV抗原表达, 降低病毒复制, 有利于逃避宿主免疫系统的监视[15-17]. 国内对于上一机制鲜有报道, 为了进一步阐述上述机制和研究HCV IRES位点功能, 我们构建了含HCV IRES的双顺反子载体.
pcDNA3, 德国Qiagen公司; Expand PCR Kit, 美国Stratgene公司; Taq 酶, 加拿大Bio-Star公司; EcoR I、 BamH I, 洛阳华美生物工程公司. HBV pcDNA3-C 和pcDNA3-S基因质粒系本室保存. 引物序列: 以下引物均在大连宝生物公司合成 HCV IRES L: 5'- GCGCGGATCCGG GCGACACTCCACCATAG -3'(nt17-36, 黑体划线为BamH-I切点)HCV IRES R: 5'-GC GAATTCGTTTTTCTTTGAGGTTTAGGATTC -3'(nt347-371, 黑体划线为EcoR I切点).
临床上已被鉴定的1例国人慢性HCV感染患者的阳性血清50 μL, 加2倍的裂解液Trazol(深圳晶美公司提供), 加样枪混匀后, 常规酚: 氯仿: 异戊醇提取, 4 °C离心13000 r/min, 14 min后取上清, 加2倍乙醇沉淀, 在离心后加750 mL/L乙醇洗涤, 沉淀烘干后加20 μL的DEPC水溶解后备用. 取含有HCV RNA的溶液, RT-PCR一步法试剂盒由深圳晶美公司提供, 按照试剂盒要求, 依次加入随机引物, HCV IRES上下游引物及含有逆转录酶的PCR扩增混合物. PCR仪为美国罗氏公司Real-time PCR仪. 反应程序为: 42 °C, 45 min; 94 °C, 5 min后进入主循环92 °C 30 s, 55 °C 30 s, 72 °C 30 s , 45个循环后收集PCR产物. PCR产物常规酚: 氯仿: 异戊醇提取后, 离心后上清直接EcoR I和BamH I双酶切, 酶切后电泳, QIAGEN(德国产)试剂盒回收PCR产物, 定向克隆入pcDNA3-S中, 所得的质粒为pcDNA3-SI. PCR和酶切鉴定后, 送入大连宝生物公司测序鉴定.
1.2.1 HBV (hepatitis B virus) 核心基因亚克隆 EcoR I酶切含有HBV 核心基因的质粒, 核心基因片段再回收. 小量提取pcDNA3-SI质粒后, EcoRI酶切后与HBV 核心基因连接, 连接后产物常规转化感受态细胞, 小量培养筛选的单菌落后提取质粒, 所得质粒称为pcDNA3-SIC. 以HCV IRES L和HCV 核心基因扩增下游引物(本室保存)作为所得质粒PCR方向鉴定引物, HBV 核心基因克隆鉴定方向后, 送大连宝生物公司测序.
1.2.2 pcDNA3-SIC在真核细胞HepG2表达 常规培养细胞, 转染前1 d将HepG2细胞接种到8孔槽中, 加入100 mL/L胎牛血清( FCS)的DMEM培养基, 在37 °C和50 mL/L的CO2中培养至细胞的融合率为50-80 %. II, 细胞转染 用QIAGENE小量提取质粒试剂盒提取pcDNA3-SIC质粒, 按lipofectamine试剂盒(boehringer mannheim biochemicals Co. USA)说明书要求, 转染HepG2细胞, PBS洗涤细胞2次后用4 °C的500 g/L甲醇和丙酮混合液, 低温固定15min, 间接免疫荧光检测.
1.2.3 免疫沉淀检测 超声波破碎转染细胞, 7 000 r/min, 离心10 min, 取上清. 取等量上清分别加入单克隆HBsAb和HBcAb, 37 °C温浴45 min后, 加入PEG6000, 终浓度为25 g/L, 4 °C过夜, 离心10000 r/min, 30 min, 收集沉淀, 30 g/L的PEG6000洗涤3次, 3 mol/L KSCN室温解离3 h, 所得样品做SDS-PAGE(分离胶为120 g/L, 浓缩胶为30 g/L, 稳流25mA, 电泳70min, 电泳仪为Bio-Rad3000Xi型), 凝胶用考马斯亮兰R-250染色.
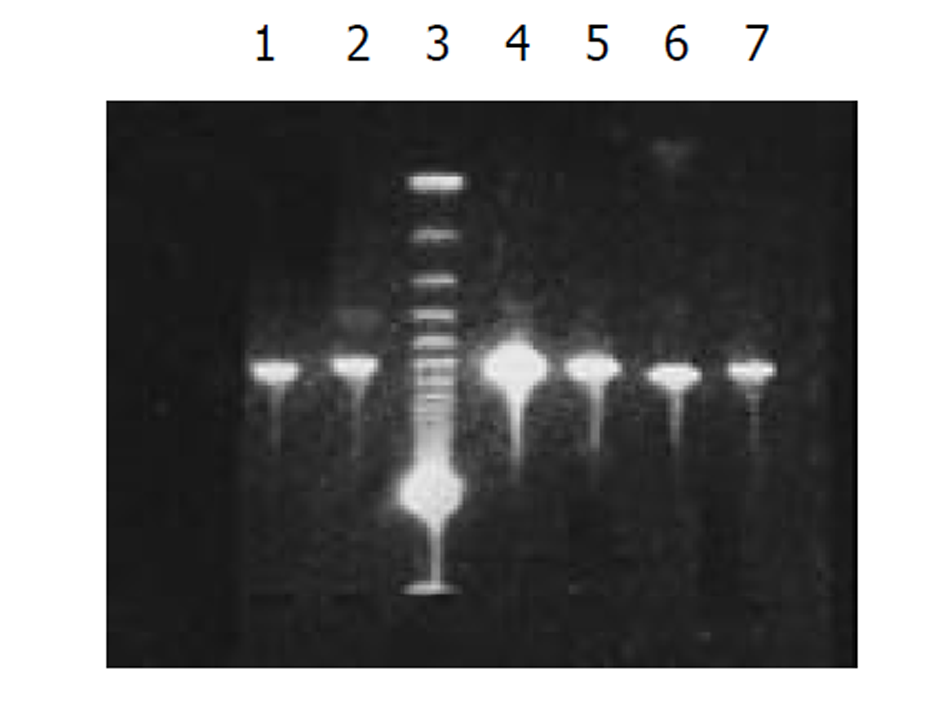
将外源性目的基因HBV S和core基因依次克隆入PCDNA3-SIC 中, PCR扩增鉴定插入PCDNA3-SIC载体中片段大小, 分别约为670 bp和550 bp, 图2.
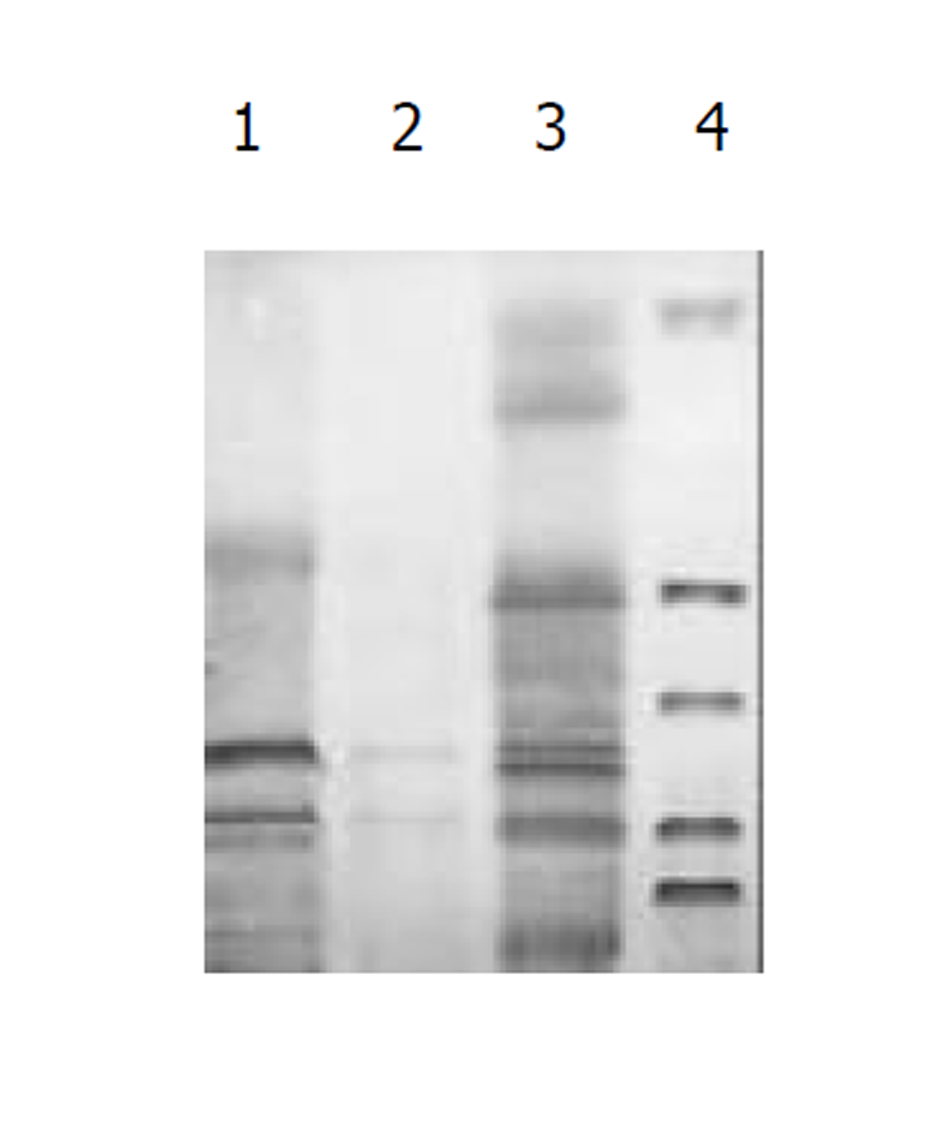
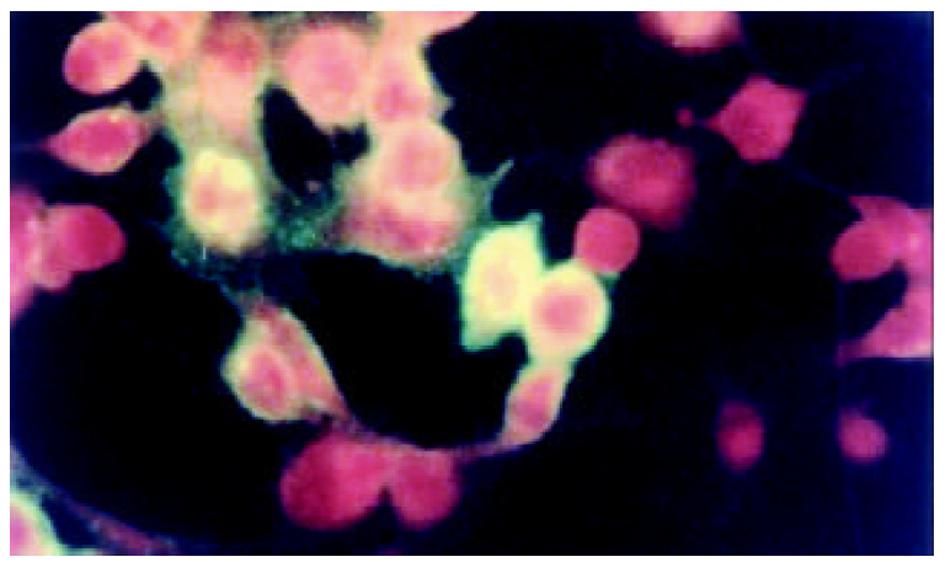
将测序和PCR鉴定完成后pcDNA3-SIC质粒, 在HepG2中转染, 间接免疫荧光染色和免疫沉淀鉴定, 图3-5.
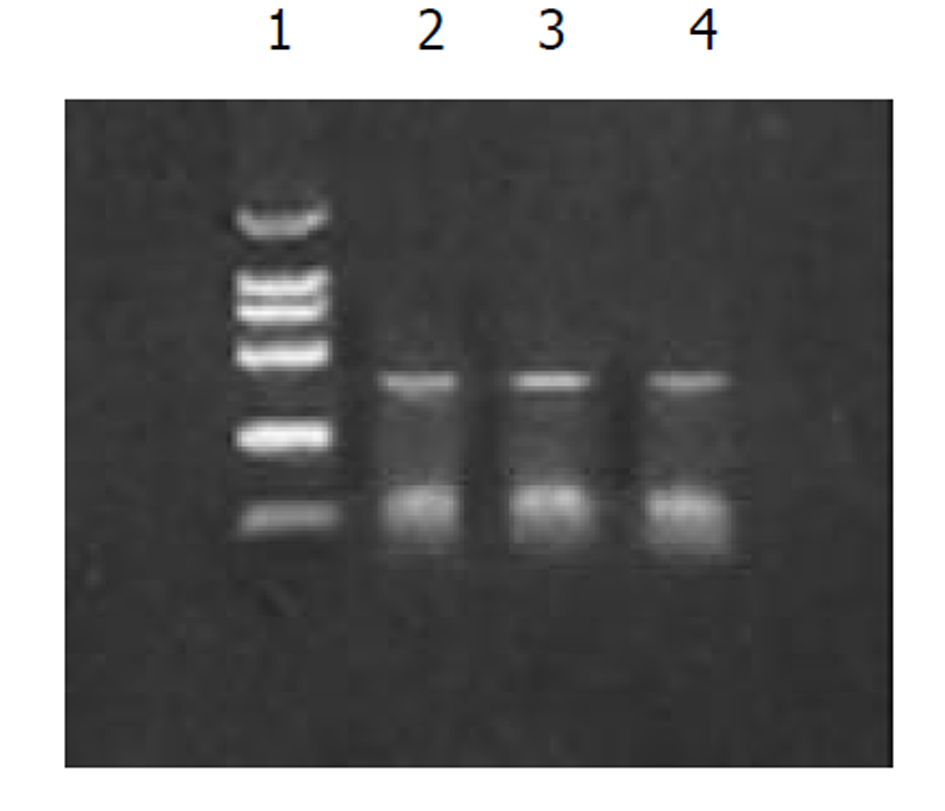
图1为HCV IRES位点RT-PCR扩增的结果, 左侧1泳道为2 kb的Mark, 右侧2、3、4泳道为PCR扩增结果, 分子大小为335 bp. 图2为PCDNA3-SIC 中HBV S和core基因扩增鉴定结果, 中间3泳道为123 bp的Mark, 左侧第一泳道为HBV core基因678 bp, 而第二泳道为HBV S基因550 bp, 4、5泳道为S, 6、7为C; 图3为pcDNA3-SIC在HepG2中转染后, 免疫沉淀鉴定结果, 最右侧4泳道为中分子质量蛋白质mark, 左侧第1泳道为 HBV S基因表达后免疫沉淀的结果, 第2泳道为无关血清对照, 第3泳道为HBV core基因表达后免疫沉淀, 可见HBV核心蛋白表达, 为2000左右.
最初描述核糖体插入位点(internal ribosome entry site, IRES)的启动帽非依赖性的翻译功能首先在细小病毒RNA的5'端非编码区[18], 随后在有多种病毒基因组中发现. 评价这些IRES元件, 特别是来源于脑心肌炎病毒(EMCV)的大约600 nt IRES序列, 可以被用做表达多种蛋白质[19-21]. 最近, HCV的基因组被证明也有一个IRES序列, 大约为200 nt左右[22-26]. 现研究表明在HCV基因组中的5'端存在一个非基因编码区(nontranslated RNA), 即5'NTR区, 长度大约300 bp, 共有4个茎环, 即I, II, III和IV. 其中第2个茎环含有IIa和IIb等2个环状结构, 第3个茎环含有5个环状结构, 即IIIa, IIIb, IIIc, IIId和IIIe, 翻译的起始位点可能位于IV环中的AUG, 而这4个茎环空间结构的正确形成对下游基因蛋白质翻译有着重要影响(见左图)[27-32]. Rijnbrand et al [33]等研究HCV IRES 功能区位于40 nt和370 nt之间, 包括一部分HCV 核心蛋白序列, 即位于茎环II和III上, 内含丰富的RNA假结, 有利于核糖体进入结合. HCV IRES能够在绝大多数细胞系中可以启动下游基因表达, 甚至包括那些非灵长类细胞, 具有帽非依赖性的强大翻译功能[34]. 也有研究表明, 茎环中第4环能够和病毒的病毒产物相结合, 对下游翻译有负调节作用, 有利于病毒的持续感染[35]. 但多数学者认为5'NTR正确的空间构像对下游基因蛋白质的翻译有重要作用.
我们通过RT-PCR扩增HCV 5'NTR部分片段(图1), 构建了pcDNA3-SIC载体, PCR鉴定(图2)和测序鉴定后, 转染真核细胞. 发现去断后的5'NTR区可完全启动下游蛋白质的表达(图4和图5), 表达后的蛋白质经免疫沉淀后, SDS-PAGE发现, 在HBcAb结合蛋白质量远大于HBsAb结合蛋白质量, IRES启动下游基因翻译不亚于CMV启动子的帽依赖结构对下游基因翻译(图3). 表明人为去断HCV 5'NTR区20个碱基, 破坏5'NTR区的第一茎环并不影响下游蛋白质表达.
在HCV基因组上的3'端同样也存在一个非编码区, 即3'NTR, 大约为40-60 nt. 在HCV RNA结构稳定性方面有重要作用[36-38]. 我们发现HCV 5'NTR区的第一个茎环在HCV蛋白质翻译并不起着重要作用, 他是否起着3'NTR相似的稳定HCV RNA作用还不得而知, 因此还有待于进一步研究. 但本研究揭示了HCV IRES位点的对下游基因翻译功能, 而且不受茎环I序列的限制, 为下一步构建含HCV IRES的双顺反子载体奠定了基础.
| 1. | Chang KM. Immunopathogenesis of hepatitis C virus infection. Clin Liver Dis. 2003;7:89-105. [DOI] |
| 2. | Chen YD, Liu MY, Yu WL, Li JQ, Dai Q, Zhou ZQ, Tisminetzky SG. Mix-infections with different genotypes of HCV and with HCV plus other hepatitis viruses in patients with hepatitis C in China. World J Gastroenterol. 2003;9:984-992. [PubMed] [DOI] |
| 3. | Roussos A, Goritsas C, Pappas T, Spanaki M, Papadaki P, Ferti A. Prevalence of hepatitis B and C markers among refugees in Athens. World J Gastroenterol. 2003;9:993-995. [PubMed] [DOI] |
| 4. | Jin J, Yang JY, Liu J, Kong YY, Wang Y, Li GD. DNA immunization with fusion genes encoding different regions of hepatitis C virus E2 fused to the gene for hepatitis B surface antigen elicits immune responses to both HCV and HBV. World J Gastroenterol. 2002;8:505-510. [DOI] |
| 5. | Tang ZY. Hepatocellular carcinoma-cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [DOI] |
| 6. | Rabe C, Pilz T, Klostermann C, Berna M, Schild HH, Sauerbruch T, Caselmann WH. Clinical characteristics and outcome of a cohort of 101 patients with hepatocellular carcinoma. World J Gastroenterol. 2001;7:208-215. [PubMed] [DOI] |
| 7. | Assy N, Paizi M, Gaitini D, Baruch Y, Spira G. Clinical implication of VEGF serum levels in cirrhotic patients with or without portal hypertension. World J Gastroenterol. 1999;5:296-300. [PubMed] |
| 8. | Agrati C, Nisii C, Oliva A, D'Offizi G, Montesano C, Pucillo LP, Poccia F. Lymphocyte distribution and intrahepatic compartmentalization during HCV infection: a main role for MHC-unrestricted T cells. Arch Immunol Ther Exp. 2002;50:307-316. |
| 9. | Lin DB, Tsai TP, Chen WK. Seroprevalence of hepatitis C virus infection and its association with natural infection of hepatitis B virus among preschool children in Taiwan. Eur J Epidemiol. 2003;18:245-249. [PubMed] [DOI] |
| 11. | Urbani S, Boni C, Missale G, Elia G, Cavallo C, Massari M, Raimondo G, Ferrari C. Virus-specific CD8+ lymphocytes share the same effector-memory phenotype but exhibit functional differences in acute hepatitis B and C. J Virol. 2002;76:12423-12434. [DOI] |
| 12. | Freeman AJ, Law MG, Kaldor JM, Dore GJ. Predicting progression to cirrhosis in chronic hepatitis C virus infection. J Viral Hepat. 2003;10:285-293. [DOI] |
| 13. | Watanabe H, Saito T, Shinzawa H, Okumoto K, Hattori E, Adachi T, Takeda T, Sugahara K, Ito JI, Saito K. Spontaneous elimination of serum hepatitis C virus (HCV) RNA in chronic HCV carriers: A population-based cohort study. J Med Virol. 2003;71:56-61. [PubMed] [DOI] |
| 14. | Kato T, Miyamoto M, Furusaka A, Date T, Yasui K, Kato J, Matsushima S, Komatsu T, Wakita T. Processing of hepatitis C virus core protein is regulated by its C-terminal sequence. J Med Virol. 2003;69:357-366. [PubMed] [DOI] |
| 15. | Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J Virol. 1999;78:9718-9725. |
| 16. | Zhang J, Yamada O, Yoshida H, Iwai T, Araki H. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology. 2002;293:841-150. [PubMed] [DOI] |
| 17. | Li D, Takyar ST, Lott WB, Gowans EJ. Amino acids 1-20 of the hepatitis C virus (HCV) core protein specifically inhibit HCV IRES-dependent translation in HepG2 cells, and inhibit both HCV IRES- and cap-dependent translation in HuH7 and CV-1 cells. J Gen Virol. 2003;84:815-825. [PubMed] [DOI] |
| 18. | Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320-325. [PubMed] [DOI] |
| 19. | Morgan RA, Couture L, Elroy-Stein O, Ragheb J, Moss B, Anderson WF. Retrovirol vectors containing patent internal ribosome entry site: development of a polycistronic gene transfer system and applications to human gene therapy. Nucl Acids Res. 1992;20:1293-1299. [DOI] |
| 20. | Gurtu V, Yan G, Zhang G. IRES bicistronic expression vectors for efficient creation of stable mammalian cell lines. Biochem Biophys Res Commun. 1996;229:295-298. [PubMed] [DOI] |
| 21. | Pu H, Cashion LM, Kretschmer PJ, Liu Z. Rapid establishment of high-producing cell lines using dicistronic vectors with glutamine synthetase as the selection marker. Mol Biotechnol. 1998;10:17-25. [PubMed] [DOI] |
| 22. | Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal ribosome entry site of hepatitis C virus RNA translation. EMBO J. 1995;14:6010-6020. [DOI] |
| 23. | Lu HH, Wimmer E. Poliovirus chimera replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci. 1996;93:1412-1417. [DOI] |
| 24. | Beales LP, Holzenburg A, Rowlands DJ. Viral internal ribosome entry site structures segregate into two distinct morphologies. J Virol. 2003;77:6574-6579. [DOI] |
| 25. | Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA-RNA interaction between the 5' nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599-606. [DOI] |
| 26. | Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J Biol Chem. 2003;278:12231-12240. [PubMed] [DOI] |
| 27. | Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5 nontranslated region of hepatitis C virus required for RNA replication. J Virol. 2001;75:12047-12057. [PubMed] [DOI] |
| 28. | Branch AD. Hepatitis C virus RNA codes for proteins and replicates: does it also trigger the interferon response? Semin Liver Dis. 2000;20:57-68. [PubMed] [DOI] |
| 29. | Hwang LH, Hsieh CL, Yen A, Chung YL, Chen DS. Involvement of the 5 proximal coding sequences of hepatitis C virus with internal initiation of viral translation. Biochem Biophys Res Commun. 1998;252:455-460. [PubMed] [DOI] |
| 30. | Frolov I, McBride MS, Rice CM. Cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5 nontranslated region chimeras. RNA. 1998;4:1418-1435. [DOI] |
| 31. | Rijnbrand RC, Abbink TE, Haasnoot PC, Spaan WJ, Bredenbeek PJ. The influence of AUG codons in the hepatitis C virus 5 nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47-56. [PubMed] [DOI] |
| 32. | Honda M, Rijnbrand R, Abell G, Kim D, Lemon SM. Natural variation in translational activities of the 5 nontranslated RNAs of hepatitis C virus gentypes Ia and Ib: evidence for a long-range RNA-RNA interaction outside of the ribosomal entry site. J Virol. 1999;73:4941-4951. [PubMed] |
| 33. | Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJ, Bredenbeek PJ. Internal entry of ribosomes is directed by 5'noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initial codon. J Virol. 1999;71:451-457. |
| 34. | Borman AM, Le Mercier P, Girard M, Kean KM. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucl Acids Res. 1997;25:925-932. [DOI] |
| 35. | Ali N, Siddiqui A. The La antigen binds 5 noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry sited-mediated translation. Proc Natl Acad Sci. 1997;94:2249-2254. [PubMed] [DOI] |
| 36. | Yi M, Lemon SM. Structure-function analysis of the 3 stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003;9:331-345. [DOI] |
| 37. | Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3 nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:53265338. [DOI] |
| 38. | Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott C, Del Vecchio AM. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3'NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457-1463. [DOI] |