Published online Jun 7, 2021. doi: 10.3748/wjg.v27.i21.2834
Peer-review started: January 23, 2021
First decision: February 10, 2021
Revised: March 30, 2021
Accepted: April 28, 2021
Article in press: April 28, 2021
Published online: June 7, 2021
Emerging evidence has demonstrated that fecal microbiota transplantation (FMT) has a promising therapeutic effect on mice with experimental colitis and patients with ulcerative colitis (UC), although the mechanism of FMT is unclear.
To evaluate the protective effect of FMT on UC and clarify its potential depen
Dextran sodium sulfate (DSS)-induced experimental colitis was established and fecal microbiota was transplanted by gavage. Severity of colon inflammation was measured by body weight, disease activity index, colon length and histological score. Gut microbiota alteration was analyzed through 16S ribosomal ribonucleic acid sequencing. The differentially expressed genes (DEGs) in the colon were obtained by transcriptome sequencing. The activation status of colonic T lymphocytes in the lamina propria was evaluated by flow cytometry.
Compared with the DSS group, the weight loss, colon length shortening and inflammation were significantly alleviated in the FMT group. The scores of disease activity index and colon histology decreased obviously after FMT. FMT restored the balance of gut microbiota, especially by upregulating the relative abundance of Lactobacillus and downregulating the relative abundance of Clostridium_sensu_stricto_1 and Turicibacter. In the transcriptomic analysis, 128 DEGs intersected after DSS treatment and FMT. Functional annotation analysis suggested that these DEGs were mainly involved in T-lymphocyte activation. In the DSS group, there was an increase in colonic T helper CD4+ and T cytotoxic CD8+ cells by flow cytometry. FMT selectively downregulated the ratio of colonic CD4+ and CD8+ T cells to maintain intestinal homeostasis. Furthermore, Clostri dium_sensu_stricto_1 was significantly related to inflammation-related genes including REG3G, CCL8 and IDO1.
FMT ameliorated DSS-induced colitis in mice via regulating the gut microbiota and T-cell modulation.
Core Tip: Previous studies shown that inflammatory bowel disease patients received satisfactory efficacy and safety after fecal microbiota transplantation (FMT) treatment. However, the mechanism of FMT remains unclear. Here, we set out animal experiments to explore the role of FMT in dextran sodium sulfate induced colitis in mice based on microbiome and transcriptome analysis.
- Citation: Wen X, Wang HG, Zhang MN, Zhang MH, Wang H, Yang XZ. Fecal microbiota transplantation ameliorates experimental colitis via gut microbiota and T-cell modulation. World J Gastroenterol 2021; 27(21): 2834-2849
- URL: https://www.wjgnet.com/1007-9327/full/v27/i21/2834.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i21.2834
Inflammatory bowel disease (IBD) is a disease of global concern with a growing prevalence in young adults. The current treatments for IBD are not completely satisfactory. Modulating microbiota to correct dysbiosis is a new approach to treating IBD. Fecal microbiota transplantation (FMT) refers to infusion of a fecal suspension prepared from a healthy donor into the gut of a patient to restore a healthy compo
FMT has shown a valid therapeutic benefit after the onset of refractory recurrent Clostridium difficile infection by reintroducing a balanced microbiota to restore the gut microbial community. FMT has emerged as a highly effective treatment for refractory Clostridium difficile infection, achieving a cure rate of over 90%[2]. Reconstructing the gut microbiota through FMT also has benefits for patients with IBD[3,4]. The diversity of microbiota with FMT treatment increases to varying degrees among relieved patients. IBD patients show a decrease in overall biodiversity of the microbial ecosystem characterized by the loss of beneficial commensal microflora and the expansion of pathogenic bacteria[5]. In our previous studies and others, IBD patients received satisfactory efficacy and safety after FMT treatment[6-8]. However, the mechanism of FMT in the treatment of IBD is still unknown. In addition to recon
This study used wild-type mice induced by dextran sodium sulfate (DSS) as a model of colitis in order to identify the role of gut microbiota in mediating the effect of FMT on DSS-induced colitis. We evaluated the protective effect of FMT on mice with DSS-induced colitis and identified the related gut microbiota and intestinal mucosal immune signaling pathway. Our findings will provide a basis for explaining the mechanism of FMT in the treatment of IBD.
C57BL/10 mice (female; 7 wk to 8 wk of age; weighing 18-20 g; specific pathogen free grade) were purchased from the Model Animal Research Center of Nanjing University. All mice were reared in the experimental animal center of the Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University in the specific pathogen free condition. Throughout the acclimatization and study periods, all animals were maintained on a 12 h light-dark cycle (21 ± 2 °C with a relatively constant humidity of 45% ± 10%) and had access to food and water ad libitum. Randomization was used to assign animals to the experimental groups. The animal experimental protocol was approved by experimental animal ethics committee of the Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, No: DW-P-2018-008-01.
To induce acute experimental colitis, mice were administered 2.5% (w/v) DSS (molecular weight, 36-50 kDa; MP Biomedicals, LLC, Irvine, CA, United States) in their drinking water and replaced it with normal water after 7 d. The mice in FMT group were fed with fecal microbiota from healthy mice from the 8th d, once every 2 d until the end of the experiment. The mice in the DSS group were fed normal saline as negative control at the same time. During intragastric administration, the procedures were strictly performed under sterile conditions. Animals were examined daily for change in net body weight and the disease activity index (DAI). The subsequent course of colitis development was evaluated daily by scoring via the DAI. DAI is a common index used to assess the severity of colitis combined with body weight loss, stool consistency and the presence of blood in the stool. The DAI score was calculated by grading on a 0-4 scale based on the following parameters: Change in bodyweight (1, 1%-5%; 2, 5%-10%; 3, 10%-15%; 4, ≥ 15%), stool consistency (0, normal; 2, loose stools; 4, diarrhea), and stool blood (0, no blood seen; 2, obvious blood with stool; 4, grossly bloody stool). At the end of the experiment, the mice were euthanized. The colon tissue was dissected; colons were measured for colon length, and tissues were examined for gross macroscopic appearance and stool consistency. Then, the colon was divided into different parts for different analysis and then fixed in 10% formalin for histological analysis or in ribonucleic acid (RNA) protective solution (Beyotime, Beijing, China) for transcriptomics analysis of colonic mucosa.
Feces were collected directly from healthy donor mice of the same strain. Fresh feces from 5 mice were pooled, mixed with sterile normal saline according to the concentration of 0.125 g/mL and homogenized immediately. The homogenate was centrifuged at 4 °C for 5 min at 1200 rpm, and the fecal sediment was used for transplantation. Gavage was carried out with a curved gavage needle appropriate for animal size. The volume of intragastric administration was 0.1 mL/10 g to each mouse, with care taken to avoid regurgitation. The two experimental groups stopped eating after 10:00 pm before transplantation, and FMT was always performed at 8:00 am.
The distal colon segments were placed in 10% neutral buffered formalin for 24 h, embedded in paraffin and cut into sections of 4 μm in thickness. The sections were then stained with hematoxylin and eosin (HE). HE-stained sections were examined for inflammation and tissue damage. Slices were evaluated by the experienced pathologist in a blinded manner, and histological score was assessed based on the following parameters according to previous research: Inflammation extent (0, none; 1, mild; 2, moderate; 3, severe), crypt aberrant (0, normal; 1, basal 1/3 damage; 2, basal 2/3 damage; 3, crypt lost and surface epithelium present; 4, crypt and surface epithelium lost), lymphocyte infiltration (0, 0%; 1, 10%; 2, 10%-25%; 3, 25%-50%; 4, > 50%) and colon wall aberrant (0, none; 1, mucosa; 2, submucosa; 3, transmural). The total histological score was the sum of all of the parameters evaluated[10].
To isolate lamina propria mononuclear cells, Peyer’s Patches were firstly removed and then colonic tissue was incubated with 5 mmol/L ethylenediamine tetraacetic acid (Biosharp, Sakai, Japan) and 1 mmol/L DTT (Biofroxx, Einhausen, Germany) at 37 °C for 30 min, followed by digestion with collagenase IV (0.3 mg/mL, Sigma, St. Louis, MO, United States), DNase I (0.25 mg/mL, Biosharp) and Dispase II (3 mg/mL, Shanghai YuanYe Biotechnology, Shanghai, China) (37 °C for 1 h). Colonic lamina propria lymphocytes were then separated with a Percoll gradient.
For immunephenotyping, mononuclear cells isolated were stained with surface markers of APC-labeled anti-mouse CD4 (Biolegend, Cat. No. 100516, San Diego, CA, United States), FITC-labeled anti-mouse CD8a (Biolegend, Cat. No. 100706) for 30 min at 4 °C in the dark. Samples were passed on a FACSCanto II flow cytometer (BD). Data were analyzed using FlowJo v10 software (FlowJo, LLC, Ashland, OR, United States).
Microbial community genomic deoxyribonucleic acid (DNA) was extracted from samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, United States) according to manufacturer’s instructions. The DNA extract was checked on 1% agarose gel, and DNA concentration and purity were determined with NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, DE, United States). The hypervariable region V3-V4 of the bacterial 16S ribosomal RNA (rRNA) gene were amplified with primer pairs 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R(5'-GGACTACHVGGGTWTCTAAT-3') by an ABI GeneAmp® 9700 PCR thermocycler (Applied Biosystems, Foster City, CA, United States). The polymerase chain reaction (PCR) amplification of 16S rRNA gene was performed as follows: Initial denaturation at 95 °C for 3 min, followed by 27 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 45 s, single extension at 72 °C for 10 min, and end at 4 °C. The PCR mixtures contain 5 × TransStart FastPfu buffer 4 μL, 2.5 mmol/L dNTPs 2 μL, forward primer (5 μmol/L) 0.8 μL, reverse primer (5 μmol/L) 0.8 μL, TransStart FastPfu DNA Polymerase 0.4 μL, template DNA 10 ng and finally ddH2O up to 20 μL. PCR reactions were performed in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to manufacturer’s instructions and quantified using Quantus™ Fluorometer (Promega, Madison, WI, United States). Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, CA, United States). The predominance of bacterial communities between groups were analyzed by linear discriminant analysis effect size method.
Total RNA from inflammatory colonic tissue was extracted. Samples about 1 cm long were for sequencing. The tissues from minimum and maximum histological score were removed. Then, we collected colon samples from four randomly chosen animals per group for sequencing, totaling 12 samples. A total amount of 2 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, United States) following manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Samples were then sequenced on an Illumina Hiseq 4000 platform, and paired-end 150 bp reads were generated. Next, we used Star and Cufflinks software to complete alignment and analyze transcripts and to make quantitative analysis of all genes. Differential genes were identified, and functional enrichment analysis of these differential genes was carried out to explore their functions. The primers were: Adapter, oligonucleotide sequences for TruSeqTM RNA and DNA Sample Prep Kits. RNA 5’Adapter (RA5), part # 15013205: 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACG ACGCTCTTCCGATCT-3’; RNA3’Adapter (RA3), part # 15013207: 5’-GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (index) ATCTCGTATGCCGTCTTCTG CTTG-3’. Gene ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GO seq R packages based Wallenius non-central hyper-geometric distribution (Young et al[11]), which can adjust for gene length bias in DEGs. Differential expression analysis was performed using the DESeq R package (1.10.1). When there was no biological repetition, TMM was firstly used to standardize the readcount data, and then DEG seq was used for difference analysis, of which the threshold was q value < 0.005 and | log2FoldChange | > 1. For differential genes, if the log2Foldchange of the gene is > 0, the differential gene was considered to be up-regulated. On the contrary, if log2Foldchange was < 0, the differential gene was considered to be down-regulated.
Metastats software was used to confirm significant differences in the relative abundance of microbiota in three groups of samples. Results were considered significant if P ≤ 0.05. Then used DEG seq for difference analysis, of which the threshold is P <0.05 and |log2FC| > 1. R psych package was used for partial Spearman correlations. r > 0.8 and P < 0.05 (strong correlation) were screened for mapping. Red edges indicate positive correlation and blue edges indicate negative correlation. The thicker the line, the stronger the correlation. The node size represents the node degree.
Differences were analyzed using a Student’s t-test or one-way analysis of variance (ANOVA) with Graphpad Prism 8.0 software (GraphPad Software Inc., La Jolla, CA, United States). Comparison among groups was made using ANOVA analysis, and comparison between two groups were made using the independent-sample t-test. Results are shown as mean ± standard error; Values of aP < 0.05, bP < 0.01, and cP < 0.001 were considered as statistically significant.
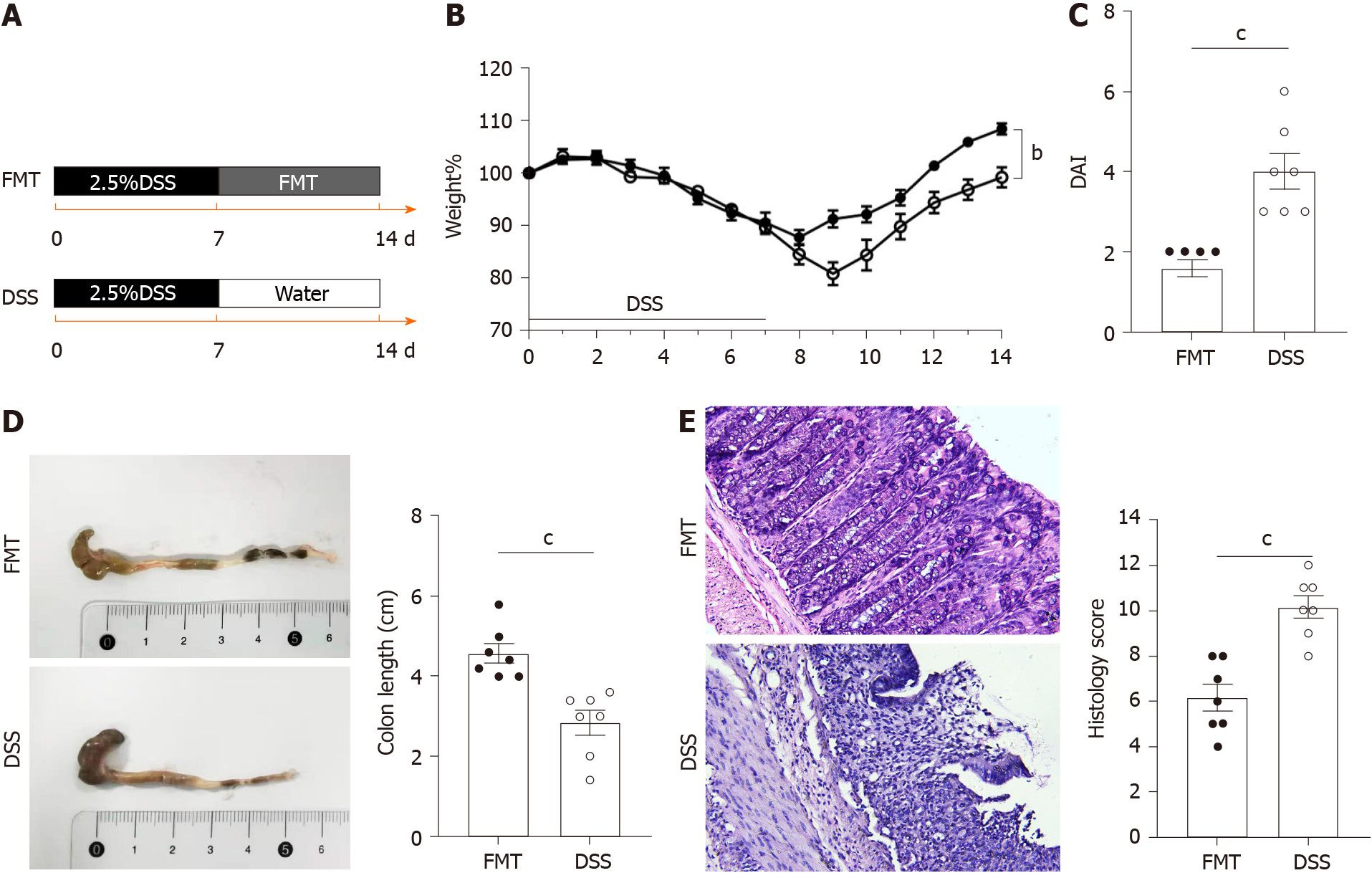
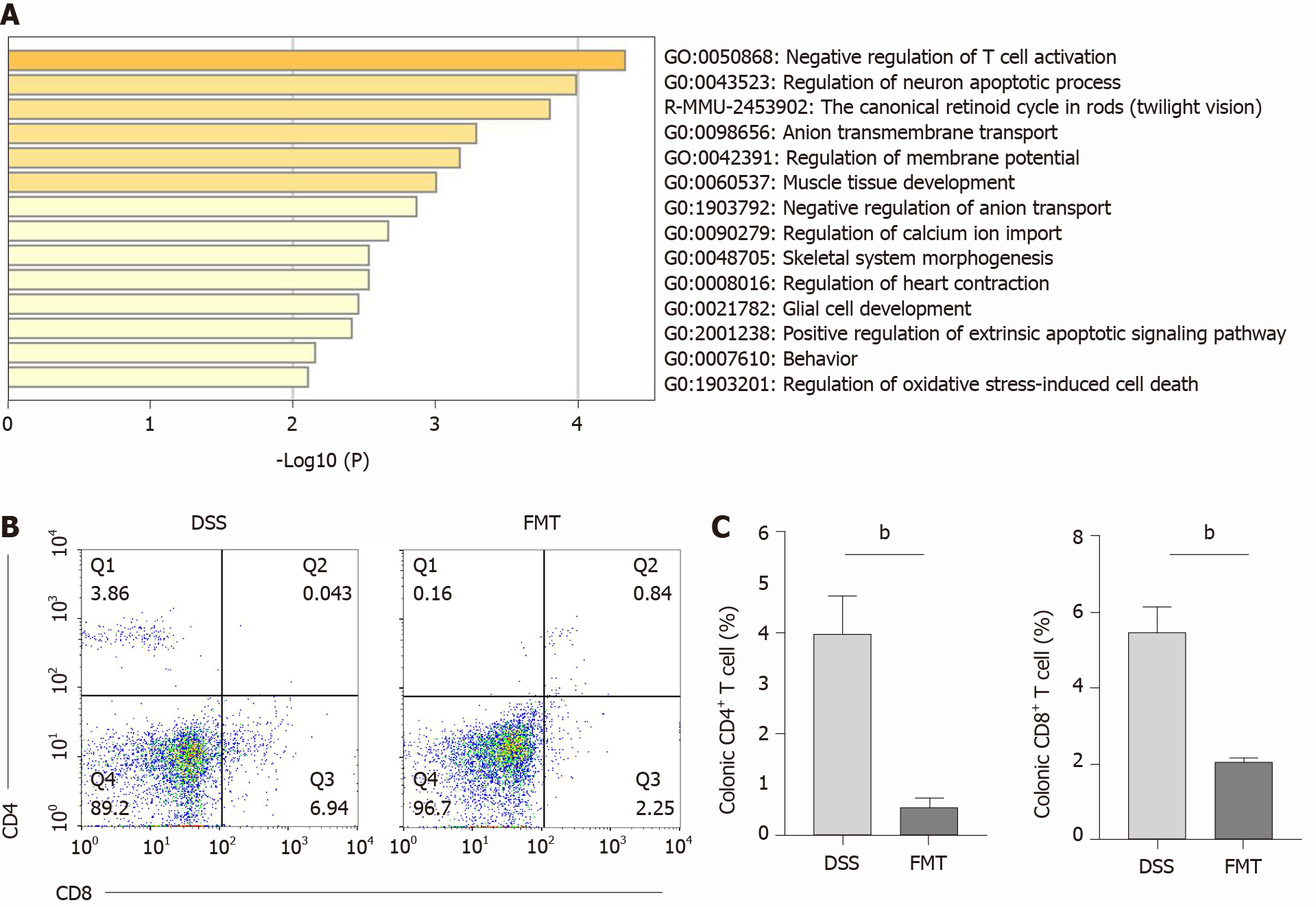
To explore the therapeutic effect of FMT on colitis, a mouse model of acute colitis induced by DSS was established. A flowchart of this study is shown in Figure 1. Five healthy mice were included as the control group. Other mice randomly drank 2.5% DSS for 7 d and were divided into the DSS and FMT groups (n = 7 each). The DSS group was gavaged with normal saline after 7 d of DSS, while the FMT group was gavaged fecal microbiota isolated from healthy mice for 7 d after DSS intervention (Figure 1A). Compared with the DSS group, FMT obviously alleviated body weight loss (bP < 0.01; Figure 1B) and DAI (cP < 0.001; Figure 1C). These findings suggested that FMT improved the clinical symptoms in DSS-induced colitis. Additionally, FMT significantly alleviated the colon shortening caused by DSS treatment (cP < 0.001; Figure 1D). To evaluate further the anti-inflammatory role of FMT, HE staining was used to assess pathological features in DSS-induced colitis. In the DSS group, colonic mucosal epithelial cells were absent, glands had lost their integrity, extensive infiltration of inflammatory cells was apparent and typical inflammatory reactions were observed. However, in the FMT group, colonic structure was relatively intact, and the histological score was lower compared with that in the DSS group (cP < 0.001; Figure 1E). These results indicated that FMT significantly attenuated the DSS-induced colonic tissue injury and inflammatory response in the mice.
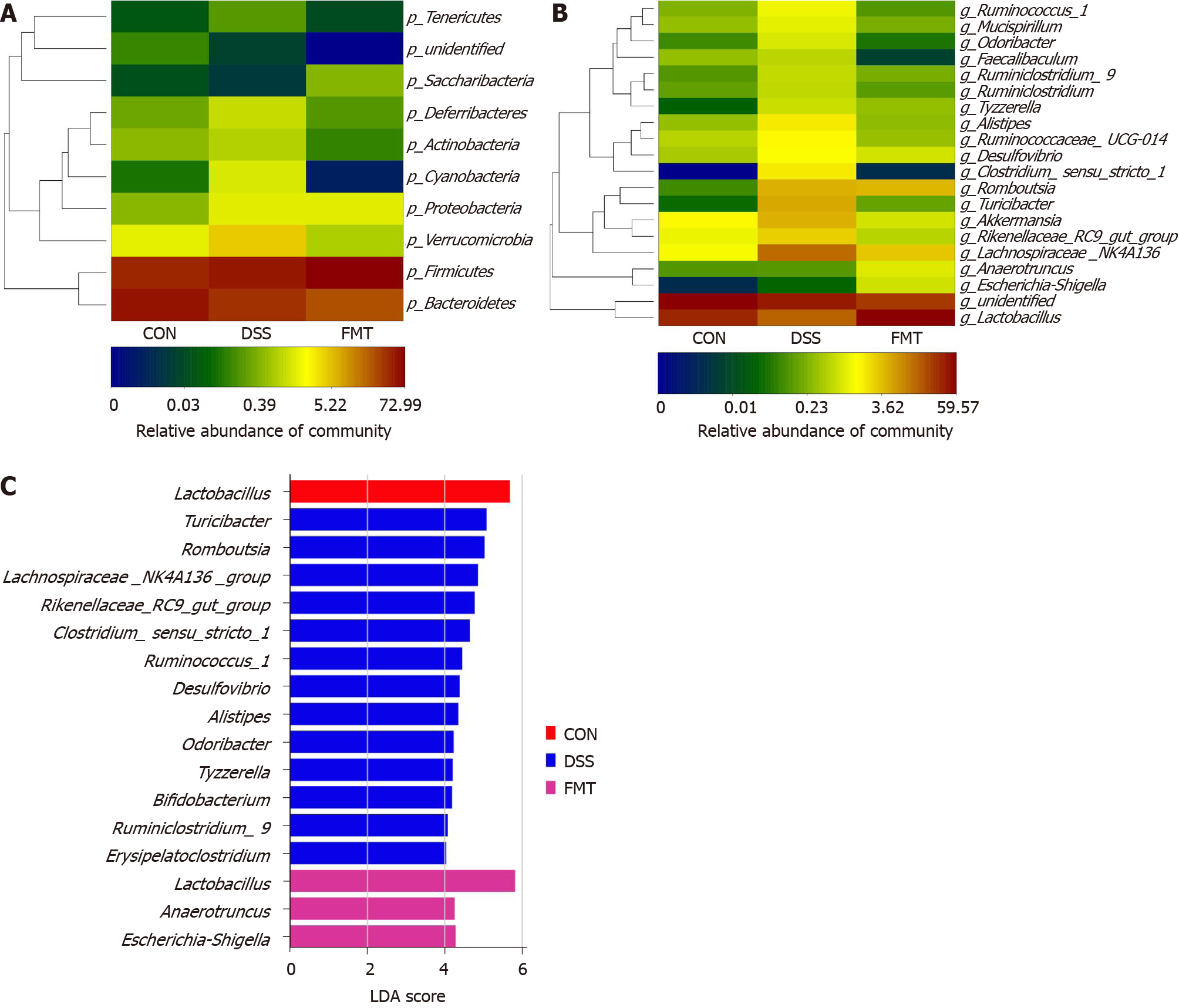
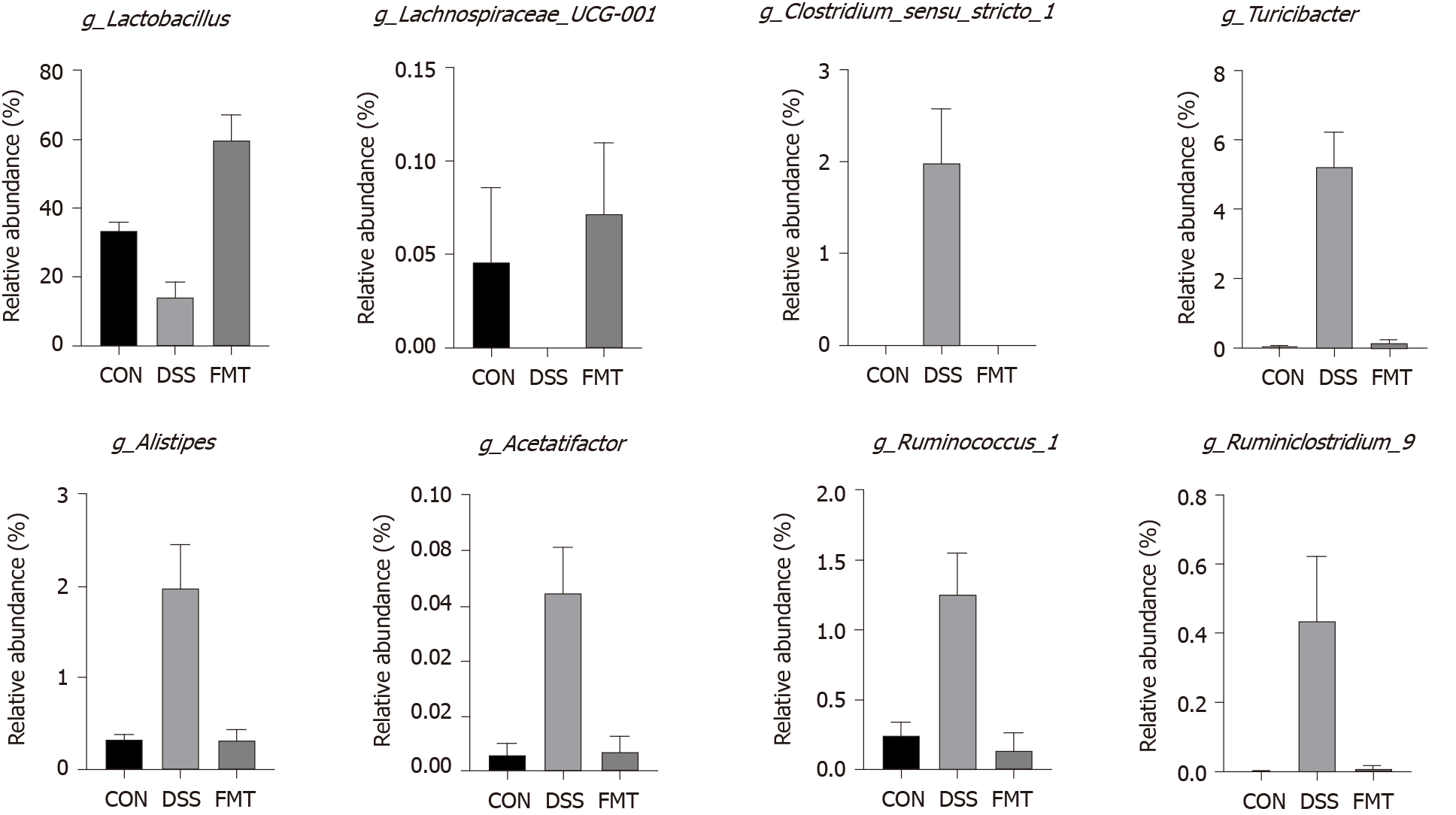
To ascertain the effects of FMT on the gut microbiota, 16S rRNA gene amplification and sequencing were conducted (Figure 2). In terms of bacterial composition at the phylum level, all samples shared similar taxonomic communities and exhibited a relatively high abundance of the phyla Bacteroidetes and Firmicutes (Figure 2A). FMT administration was associated with variations in the abundance of specific taxa at the genus level (Figure 2B). The linear discriminant analysis effect size analysis showed that Lactobacillales was enriched in the gut of untreated mice, while Clostridiales was enriched in DSS-induced colitis mice. After FMT, Lactobacillales became obviously enriched, compared with the DSS group (Figure 2C). At the genus level, Clostridium_sensu_ stricto_1 (cP = 0.001, one-way ANOVA), Turicibacter (cP < 0.001, one-way ANOVA), Alistipes (cP = 0.001, one-way ANOVA), Acetatifactor (cP = 0.001, one-way ANOVA), Ruminococcus_1 (bP < 0.01, one-way ANOVA) and Ruminiclostridium_6 (aP < 0.05, one-way ANOVA), which were expanded in colitis, were reduced after FMT and returned to being comparable to those observed in healthy mice. On the contrary, Lactobacillus (cP < 0.001, one-way ANOVA) and Lachnospiraceae_UCG-001 (aP < 0.05, one-way ANOVA) displayed a trend of high abundance in the FMT group (Figure 3), and there was no significant difference in Lactobacillus between control and transplanted mice. Collectively, these results showed that FMT significantly improved the imbalanced gut microbiota induced by DSS in mice.
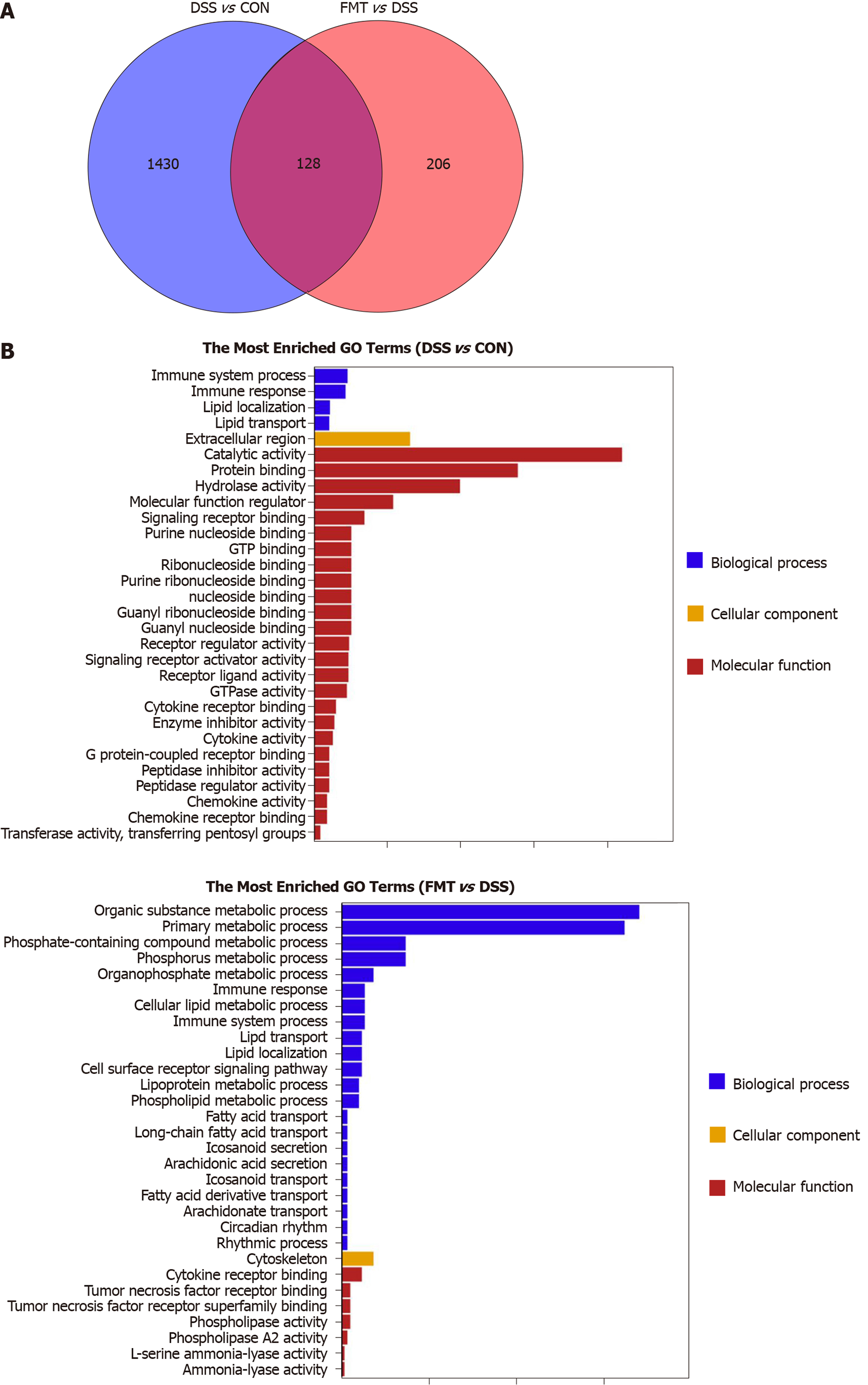
To investigate the molecular target of FMT, we used RNA sequencing in colonic tissue samples from control, DSS and FMT groups. We found that 1558 genes were differentially expressed in the DSS group compared to the control group, and 334 genes were differentially expressed in the FMT compared to the DSS group (Figure 4A). The GO enrichment histogram of DEGs showed the biological process, cellular component and molecular function. The bar chart shows the top 30 GO terms with the most significant enrichment (Figure 4B). The most enriched GO terms were immune system process and metabolic process. Additionally, there were 128 DEGs in the DSS group compared with the control and FMT groups. GO analysis showed that these genes mainly participated in T-cell activation induced by DSS and inhibited by FMT (Figure 5A). T-cell activation status was determined by surface marker staining, using flow cytometry. As expected, T-cell populations isolated from colons of FMT-treated mice showed reduced activity as compared to those isolated from colitis mice that received DSS (Figure 5B, C, bP < 0.01). Phenotypically, mice receiving DSS displayed a higher proportion of CD4+ and CD8+ T cells. However, there was a significant decrease in CD4+ and CD8+ T cells in colons following FMT, supporting the observation that colonic T cells in FMT-treated mice manifested decreased activation. These results indicated that FMT alleviated DSS-induced colitis by inhibiting intestinal immunity related to T-cell modulation.
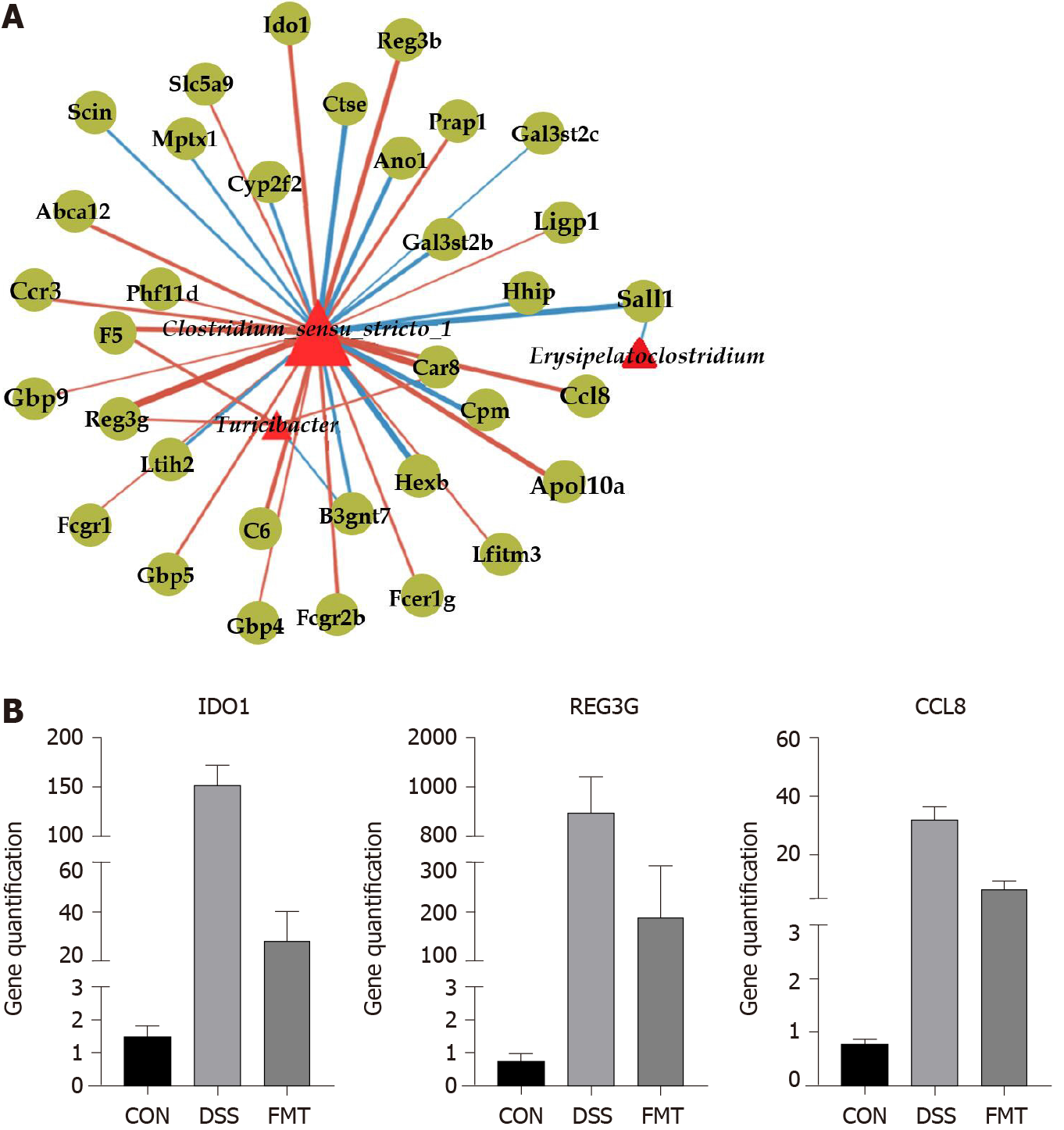
The process by which the gut microbiota regulates intestinal immunity is complicated. The gut microbiota and T-cell immunity were both involved in the biological process of FMT treatment of colitis. Based on this, we tried to find the potential gut microbiota/immune axis-mediated signaling pathway. The correlation analysis between gut microbiota and transcriptome showed that Clostridium_sensu_stricto_1 was the most enriched genus (Figure 6A). The DEGs related to gut microbiota are listed in Table 1. The DEGs, such as IDO1, CCL8 and REG3G, were positively correlated with the relative abundance of Clostridium_sensu_stricto_1 (bP < 0.01). The DEGs such as REG3G were positively correlated with the relative abundance of Turicibacter (bP < 0.01). Relative quantification of transcript level is shown in Figure 6B (all cP < 0.001, one-way ANOVA). These results provide a potential Clostri dium_ sensu_stricto_1-mediated immune regulation for FMT to treat colitis.
| Number | Gene name | Target microbiota | r value | P value |
| 1 | Ccl8 | Clostridium_sensu_stricto_1 | 0.87 | < 0.001 |
| 2 | C6 | Clostridium_sensu_stricto_1 | 0.87 | < 0.001 |
| 3 | Ido1 | Clostridium_sensu_stricto_1 | 0.87 | < 0.001 |
| 4 | Abca12 | Clostridium_sensu_stricto_1 | 0.85 | < 0.001 |
| 5 | Fcgr2b | Clostridium_sensu_stricto_1 | 0.84 | < 0.001 |
| 6 | Prap1 | Clostridium_sensu_stricto_1 | 0.84 | < 0.001 |
| 7 | F5 | Turicibacter | 0.84 | < 0.001 |
| 8 | Ccr3 | Clostridium_sensu_stricto_1 | 0.84 | < 0.001 |
| 9 | Fcer1g | Clostridium_sensu_stricto_1 | 0.83 | < 0.001 |
| 10 | Gbp5 | Clostridium_sensu_stricto_1 | 0.83 | < 0.001 |
| 11 | Slc5a9 | Clostridium_sensu_stricto_1 | 0.82 | = 0.001 |
| 12 | Reg3g | Turicibacter | 0.82 | = 0.001 |
| 13 | Gbp4 | Clostridium_sensu_stricto_1 | 0.82 | = 0.001 |
| 14 | Ifitm3 | Clostridium_sensu_stricto_1 | 0.82 | = 0.001 |
| 15 | Car8 | Turicibacter | 0.81 | = 0.001 |
| 16 | Phf11d | Clostridium_sensu_stricto_1 | 0.81 | = 0.001 |
| 17 | Fcgr1 | Clostridium_sensu_stricto_1 | 0.8 | = 0.002 |
| 18 | Gbp9 | Clostridium_sensu_stricto_1 | 0.8 | = 0.002 |
| 19 | Iigp1 | Clostridium_sensu_stricto_1 | 0.8 | = 0.002 |
| 20 | Gal3st2c | Clostridium_sensu_stricto_1 | -0.8 | = 0.002 |
| 21 | Sall1 | Erysipelatoclostridium | -0.81 | = 0.001 |
| 22 | B3gnt7 | Turicibacter | -0.81 | = 0.001 |
| 23 | Mptx1 | Clostridium_sensu_stricto_1 | -0.83 | < 0.001 |
| 24 | Scin | Clostridium_sensu_stricto_1 | -0.83 | < 0.001 |
| 25 | Itih2 | Clostridium_sensu_stricto_1 | -0.84 | < 0.001 |
| 26 | Cyp2f2 | Clostridium_sensu_stricto_1 | -0.84 | < 0.001 |
| 27 | B3gnt7 | Clostridium_sensu_stricto_1 | -0.85 | < 0.001 |
| 28 | Hhip | Clostridium_sensu_stricto_1 | -0.85 | < 0.001 |
| 29 | Gal3st2b | Clostridium_sensu_stricto_1 | -0.86 | < 0.001 |
| 30 | Ano1 | Clostridium_sensu_stricto_1 | -0.87 | < 0.001 |
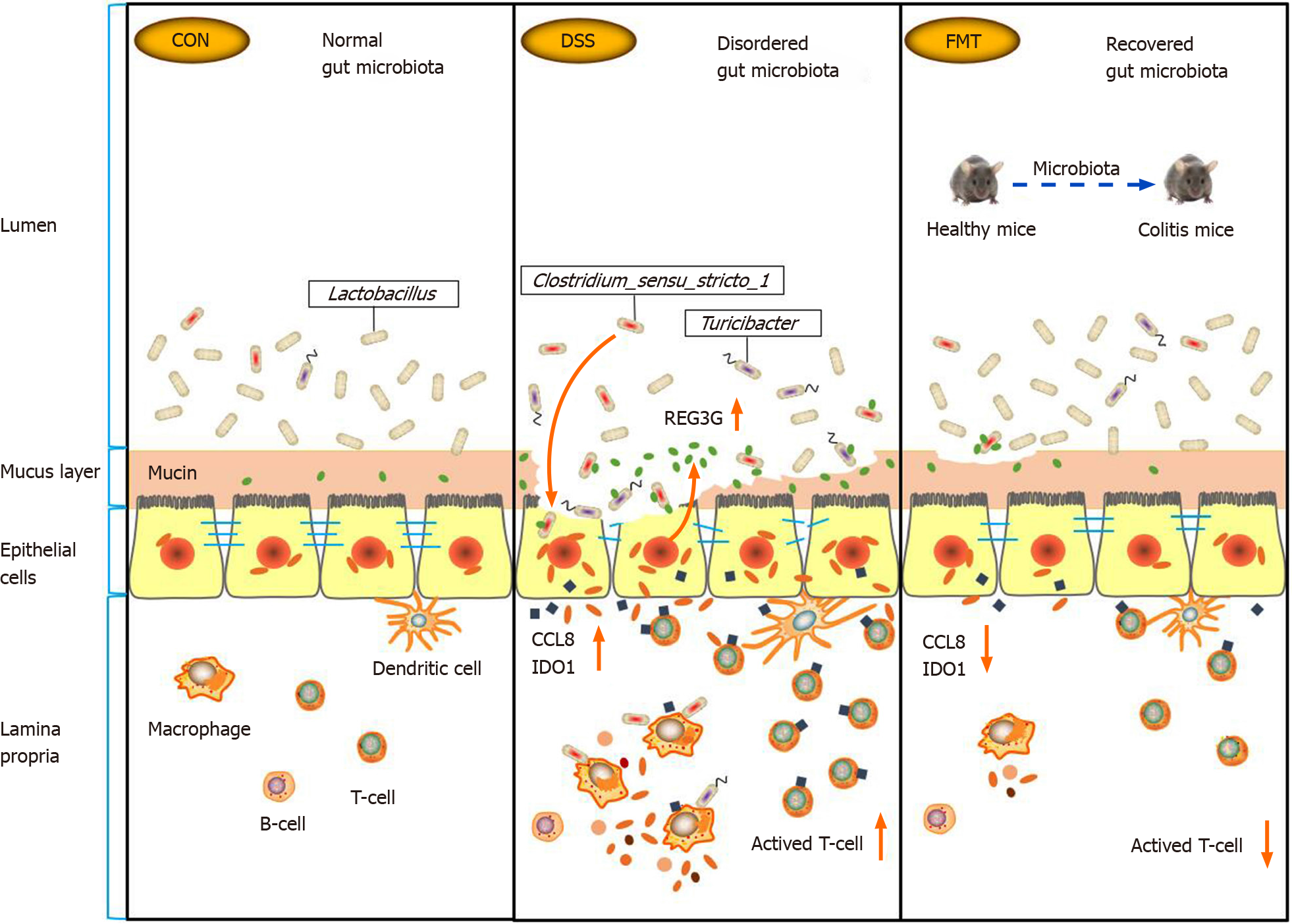
FMT significantly modulated the dysbacteriosis induced by DSS and restored the disordered gut microbiota to normal level. Meanwhile, FMT protected intestinal epithelial barrier disruption induced by DSS via upregulating the relative abundance of Lactobacillus and downregulating the relative abundance of Clos tridium_sensu_stricto_1 and Turicibacter. Furthermore, after FMT treatment, the activation of T lymphocytes was significantly inhibited (Figure 7). Hypothetically, FMT may improve experimental colitis by increasing relative abundance of Lactobacillus and regulating T-cell function.
FMT has been shown to be effective in treating ulcerative colitis (UC), but its mechanism is still unclear. In this study, 16s rRNA and transcriptome sequencing were used to prove that FMT alleviated DSS-induced colitis in mice by improving the gut microbiota and regulating T-cell function.
Lactobacillus is considered a genus of beneficial bacteria in the gut and is abundant in healthy individuals. During the inflammation stage in the colon, the abundance of Lactobacillus decreased significantly[12]. Our data suggested that Lactobacillus was the main genus in healthy mice, but its abundance decreased significantly in DSS-induced colitis. However, this process was blocked by FMT. After transplanting the resident microbiota to colitis mice, the abundance of Lactobacillus increased significantly and it became the dominant genus in the FMT group. Similarly, an increase in Lactobacillus was observed after FMT treatment in UC patients[13]. Supplementing Lactobacillus significantly prevents colonic shortening and has a therapeutic effect on DSS-induced colitis[14]. The underlying mechanism of Lactobacillus in colitis is that it promotes beneficial metabolic pathways[15] that inhibit group 3 innate lymphoid cells[16], increasing induction of regulatory T lymphocytes[17,18], decreasing inflammatory pathways[19] and strengthening gut barrier function[20]. In UC mice, Lactobacillus induced regulatory T (Treg) cell differentiation and had an impact on intestinal expression of antimicrobial peptides[21]. In detail, the Treg cells expand with decreasing levels of adenosine monophosphate in colonic tissues and thus ameliorate UC in the mice[17]. Lactobacillus also promotes the release of anti-inflammatory interleukin (IL)-10 by T cells, which is pivotal for the resolution of intestinal inflammation[22,23] by reducing the proliferative capacity of proinflammatory T cells[24]. It should be noted that not all bacterial strains of Lactobacillus exert an immunoregulatory action on T cells[21]. Thus, the role of Lactobacillus in the pathogenesis of intestinal inflammation requires further study. Besides, Clostridium_ sensu_stricto_1[25] and Turicibacter[26] are deemed to be opportunistic pathogens associated with colitis.
In our study, FMT significantly reduced the relative abundance of Clostridi um_sensu_stricto_1, Turicibacter and Ruminococcus, which were increased after DSS treatment. In IBD patients, the abundance of these opportunistic pathogens also increased[27]. Turicibacter positively correlated with proinflammatory cytokines, such as IL-6, IL-1β, tumor necrosis factor-α and interferon-γ[28]. Turicibacter exhibited higher relative abundance in IBD patients and mouse models, further inducing more severe colitis[29,30]. As reported, Turicibacter was significantly less abundant in diarrhea-dominant irritable bowel syndrome patients[31]. Recent studies also indicate an beneficial role for Turicibacter in host serotonin metabolism[32]. These findings show the complex role of Turicibacter in chronic inflammatory settings. As recently reported, Ruminococcus may drive high neutrophil-to-lymphocyte ratios that are broadly characteristic of poor disease outcomes in IBDs[33]. Ruminococcus affects the efficacy of FMT, because the presence of higher levels of Ruminococcus in donors makes FMT more likely to fail[34]. To our knowledge, the relative abundance of Clostridium_sensu_stricto_1, Turicibacter and Ruminococcus was significantly decreased after FMT. Therefore, FMT may regulate the gut microbiota to exert an anti-inflammatory effect in colitis.
In UC, the balance of commensal microbes and the immune system is impaired. In IBD patients, CD4+ T cells may exhibit significantly high pathogenicity, characterized by enhanced T-cell activation[35]. The reactivity to intestinal bacteria is a normal property of the human CD4+ T-cell repertoire and does not necessarily indicate disrupted interactions between immune cells and the commensal microbiota[36]. Drugs commonly used in the treatment of IBD, such as anti-tumor necrosis factor-α agents, cyclosporin A and azathioprine, have been demonstrated to induce apoptosis of lamina propria T cells in IBD[37]. Therefore, targeting T cells shows potential therapeutic effects. In fact, FMT regulates intestinal immunity. Therapeutic FMT administration during experimental colitis reduces colonic inflammation and initiates the restoration of intestinal homeostasis through regulating innate and adaptive immune responses in the intestinal mucosa[24]. In this work, a total of 128 DEGs were distinguished by comparing the control, DSS and FMT groups. Functional annotation analysis showed that these DEGs are mainly involved in the activation of T lymphocytes. After FMT, the activation of T lymphocytes was significantly inhibited and indoleamine 2,3-dioxygenase 1 (IDO1) was mainly involved in this process. In IBD patients and experimental colitis, overexpression of IDO1 had a positive correlation with the severity of the disease[38,39]. Likewise, there is a higher presence of IDO-producing DCs and Tregs in the lamina propria of IBD patients compared with healthy subjects, which may represent a role of regulating inflammatory me-chanism[40]. Pharmacological inhibition of IDO1 alleviates DSS-induced colitis[39,41]. In a recent study, gut microbiota from IDO1-knockout mice could directly attenuate the severity of DSS-induced colitis[42]. These results suggested that IDO1 regulated the gut microbiota to maintain intestinal homeostasis in colitis. According to our results, IDO1 expression was increased by DSS but was significantly inhibited after FMT at the transcriptional level. The reduction of intestinal T-lymphocyte activation indicates reduction of intestinal pathogenic bacteria. This can explain how the gut microbiota improved after FMT, thereby reducing activation of T cells.
In this study, we investigated the relationship between the gut microbiota and colon transcriptome in colitis. There was a significant positive correlation between Clostridium_sensu_stricto_1 and expression of antimicrobial C-type lectin regenerating islet-derived 3 gamma (REG3G), monocyte chemoattractant protein 2 (CCL8) and IDO1. REG3G belongs to the C-type lectin antimicrobial peptide family, which is closely regulated by T helper 17/IL-17A and prevents pathogenic bacteria from invading the colonic mucosa[43]. As previously described, the impact of Lactobacillus on antimicrobial peptides relied on the presence of the gut microbiota[21]. It is a defense mechanism widely produced by intestinal epithelial cells to regulate the dynamic balance between microbiota and host epithelium[44]. Colonic macrophages act as a major source of CCL8 chemokines that trigger further recruitment of their proinflammatory monocyte precursors[45]. After administering lipopolysaccharide, there was a marked increase in the expression of the monocyte chemoattractant CCL8[46]. As in animal colitis, CCL8 is significantly increased in active compared with quiescent IBD[45,47]. In our study, these highly expressed genes were positively correlated with Clostridium_sensu_stricto_1 in DSS-induced colitis, but the expression was significantly lower in the FMT group. These results indicated that FMT alleviated mouse colitis via downregulation of the expression of these microbiota-related genes.
The current study had some limitations. Firstly, the current 16S sequencing mainly included bacterial and archaeal genomes, ignoring nonbacterial organisms, such as viruses, fungi and phages. Secondly, animal studies have some limitations in evaluating the mechanism of FMT. Administration of DSS to induce subclinical colitis, while convenient, does not yield a model that perfectly mimics the pathogenesis of IBD, since DSS causes a chemical colitis through epithelial injury. Thirdly, we analyzed the correlation between gut microbiota and gene expression, but the causal relationship was not further explained. In this study, the regulation of gut microbiota and colon transcriptome by FMT may be the key to the anti-inflammatory role in colitis. We will carry out clinical studies to verify the changes in gut microbiota and transcriptome in FMT treatment of UC.
In this study, we explored the mechanism of FMT in the treatment of experimental colitis by analysis of gut microbiota and colonic transcriptome. FMT contributed to alleviation of murine colitis with increased abundance of Lactobacillus and decreased Clostridium_sensu_stricto_1 and Turicibacter. In transcriptomics, FMT inhibited activation of T lymphocytes in the colon by regulating gene expression related to the microbiota. Therefore, we propose that FMT regulates the gut microbiota and T lymphocytes function, and thereby alleviates colitis. The causal relationship between the gut microbiota and UC needs to be further studied.
Ulcerative colitis (UC) is a growing global disease in which gut microbiota dysbiosis plays an important pathogenic role. However, the current drugs for UC treatment are far from optimal. Therefore, alternative safe and effective new treatments need to be developed.
Clinical practice has confirmed the therapeutic value of fecal microbiota transplanta
The aim of the study was to explore the effect and mechanism of FMT on regulating the balance of gut microbiota and anti-inflammation in dextran sulfate sodium (DSS)-induced colitis.
Experimental colitis was induced by DSS, then the severity of intestinal inflammation was evaluated by body weight, colon length, disease activity index and histological scores. Gut microbiota alteration was analyzed through 16S rRNA sequencing. Transcriptome sequencing was used to screen differentially expressed genes in colon. The frequency of immune cells in lamina propria were phenotyped by flow cytometry.
DSS-induced weight loss, colon length shortening, disease activity index score and histological score were significantly alleviated after FMT treatment. 16S rRNA sequencing indicated that FMT up-regulated the relative abundance of Lactobacillus and down-regulated the relative abundance of Clostridium_sensu_stricto_1 and Turicibacter. Transcriptomics-based differential gene analysis showed that FMT could regulate colonic T cell function. Further flow cytometry analysis showed that FMT downregulated the number of colonic CD4+ and CD8+ T cells to maintain intestinal homeostasis. Moreover, we found that the abundance of Clostridium_sensu_stricto_1 was positively correlated with the expression of inflammation-related genes such as REG3G, CCL8 and IDO1.
FMT alleviated DSS-induced colitis in mice by improving the gut microbiota and regulating T-cell function.
This study has initially revealed the mechanism of FMT in the treatment of colitis, which will be confirmed by human studies in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Barisani D, Sitkin S, Snyder AM, Sun C S-Editor: Liu M L-Editor: Filipodia P-Editor: Liu JH
| 1. | Zhang T, Lu G, Zhao Z, Liu Y, Shen Q, Li P, Chen Y, Yin H, Wang H, Marcella C, Cui B, Cheng L, Ji G, Zhang F. Washed microbiota transplantation vs. manual fecal microbiota transplantation: clinical findings, animal studies and in vitro screening. Protein Cell. 2020;11:251-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 2. | Tariq R, Disbrow MB, Dibaise JK, Orenstein R, Saha S, Solanky D, Loftus EV, Pardi DS, Khanna S. Efficacy of Fecal Microbiota Transplantation for Recurrent C. Difficile Infection in Inflammatory Bowel Disease. Inflamm Bowel Dis. 2020;26:1415-1420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 3. | Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, Rosewarne CP, Bickley C, Peters C, Schoeman MN, Conlon MA, Roberts-Thomson IC, Andrews JM. Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial. JAMA. 2019;321:156-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 414] [Cited by in F6Publishing: 481] [Article Influence: 96.2] [Reference Citation Analysis (0)] |
| 4. | Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, Xuan W, Lin E, Mitchell HM, Borody TJ. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218-1228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 710] [Cited by in F6Publishing: 752] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 5. | Yalchin M, Segal JP, Mullish BH, Quraishi MN, Iqbal TH, Marchesi JR, Hart AL. Gaps in knowledge and future directions for the use of faecal microbiota transplant in the treatment of inflammatory bowel disease. Therap Adv Gastroenterol. 2019;12:1756284819891038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Wang H, Cui B, Li Q, Ding X, Li P, Zhang T, Yang X, Ji G, Zhang F. The Safety of Fecal Microbiota Transplantation for Crohn's Disease: Findings from A Long-Term Study. Adv Ther. 2018;35:1935-1944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 7. | Li Q, Ding X, Liu K, Marcella C, Liu X, Zhang T, Liu Y, Li P, Xiang L, Cui B, Wang J, Bai J, Zhang F. Fecal Microbiota Transplantation for Ulcerative Colitis: The Optimum Timing and Gut Microbiota as Predictors for Long-Term Clinical Outcomes. Clin Transl Gastroenterol. 2020;11:e00224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 8. | Li Q, Zhang T, Ding X, Xiang L, Cui B, Buch H, Zhang F. Enhancing patient adherence to fecal microbiota transplantation maintains the long-term clinical effects in ulcerative colitis. Eur J Gastroenterol Hepatol. 2020;32:955-962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Burrello C, Giuffrè MR, Macandog AD, Diaz-Basabe A, Cribiù FM, Lopez G, Borgo F, Nezi L, Caprioli F, Vecchi M, Facciotti F. Fecal Microbiota Transplantation Controls Murine Chronic Intestinal Inflammation by Modulating Immune Cell Functions and Gut Microbiota Composition. Cells. 2019;8:517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 10. | Li C, Ai G, Wang Y, Lu Q, Luo C, Tan L, Lin G, Liu Y, Li Y, Zeng H, Chen J, Lin Z, Xian Y, Huang X, Xie J, Su Z. Oxyberberine, a novel gut microbiota-mediated metabolite of berberine, possesses superior anti-colitis effect: Impact on intestinal epithelial barrier, gut microbiota profile and TLR4-MyD88-NF-κB pathway. Pharmacol Res. 2020;152:104603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 11. | Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3989] [Cited by in F6Publishing: 4212] [Article Influence: 300.9] [Reference Citation Analysis (0)] |
| 12. | Miranda PM, De Palma G, Serkis V, Lu J, Louis-Auguste MP, McCarville JL, Verdu EF, Collins SM, Bercik P. High salt diet exacerbates colitis in mice by decreasing Lactobacillus levels and butyrate production. Microbiome. 2018;6:57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 150] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 13. | Mańkowska-Wierzbicka D, Stelmach-Mardas M, Gabryel M, Tomczak H, Skrzypczak-Zielińska M, Zakerska-Banaszak O, Sowińska A, Mahadea D, Baturo A, Wolko Ł, Słomski R, Dobrowolska A. The Effectiveness of Multi-Session FMT Treatment in Active Ulcerative Colitis Patients: A Pilot Study. Biomedicines. 2020;8:268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Hasannejad-Bibalan M, Mojtahedi A, Eshaghi M, Rohani M, Pourshafie MR, Talebi M. The effect of selected Lactobacillus strains on dextran sulfate sodium-induced mouse colitis model. Acta Microbiol Immunol Hung. 2020;67:138-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Tamura K, Sasaki H, Shiga K, Miyakawa H, Shibata S. The Timing Effects of Soy Protein Intake on Mice Gut Microbiota. Nutrients. 2019;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Wang T, Zheng N, Luo Q, Jiang L, He B, Yuan X, Shen L. Probiotics Lactobacillus reuteri Abrogates Immune Checkpoint Blockade-Associated Colitis by Inhibiting Group 3 Innate Lymphoid Cells. Front Immunol. 2019;10:1235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 17. | Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, Hattori M, Takeshita K, Kanai T, Saijo S, Ohno N, Iwakura Y. Inhibition of Dectin-1 Signaling Ameliorates Colitis by Inducing Lactobacillus-Mediated Regulatory T Cell Expansion in the Intestine. Cell Host Microbe. 2015;18:183-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 18. | Kim DH, Kim S, Ahn JB, Kim JH, Ma HW, Seo DH, Che X, Park KC, Jeon JY, Kim SY, Lee HC, Lee JY, Kim TI, Kim WH, Kim SW, Cheon JH. Lactobacillus plantarum CBT LP3 ameliorates colitis via modulating T cells in mice. Int J Med Microbiol. 2020;310:151391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 19. | El-Baz AM, Khodir AE, Adel El-Sokkary MM, Shata A. The protective effect of Lactobacillus vs 5-aminosalicylic acid in ulcerative colitis model by modulation of gut microbiota and Nrf2/Ho-1 pathway. Life Sci. 2020;256:117927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 20. | Wang Y, Gu Y, Fang K, Mao K, Dou J, Fan H, Zhou C, Wang H. Lactobacillus acidophilus and Clostridium butyricum ameliorate colitis in murine by strengthening the gut barrier function and decreasing inflammatory factors. Benef Microbes. 2018;9:775-787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | Hrdý J, Alard J, Couturier-Maillard A, Boulard O, Boutillier D, Delacre M, Lapadatescu C, Cesaro A, Blanc P, Pot B, Ryffel B, Chamaillard M, Grangette C. Lactobacillus reuteri 5454 and Bifidobacterium animalis ssp. lactis 5764 improve colitis while differentially impacting dendritic cells maturation and antimicrobial responses. Sci Rep. 2020;10:5345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 22. | Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. 2005;174:3237-3246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 382] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 23. | Jang YJ, Kim WK, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes. 2019;10:696-711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 24. | Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J, Colucci A, Guglietta S, Carloni S, Guglielmetti S, Taverniti V, Nizzoli G, Bosari S, Caprioli F, Rescigno M, Facciotti F. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018;9:5184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 25. | Zou J, Shen Y, Chen M, Zhang Z, Xiao S, Liu C, Wan Y, Yang L, Jiang S, Shang E, Qian D, Duan J. Lizhong decoction ameliorates ulcerative colitis in mice via modulating gut microbiota and its metabolites. Appl Microbiol Biotechnol. 2020;104:5999-6012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 26. | Wan P, Peng Y, Chen G, Xie M, Dai Z, Huang K, Dong W, Zeng X, Sun Y. Modulation of gut microbiota by Ilex kudingcha improves dextran sulfate sodium-induced colitis. Food Res Int. 2019;126:108595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 27. | Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, Xavier RJ. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293-305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 888] [Cited by in F6Publishing: 881] [Article Influence: 176.2] [Reference Citation Analysis (0)] |
| 28. | Liang YN, Yu JG, Zhang DB, Zhang Z, Ren LL, Li LH, Wang Z, Tang ZS. Indigo Naturalis Ameliorates Dextran Sulfate Sodium-Induced Colitis in Mice by Modulating the Intestinal Microbiota Community. Molecules. 2019;24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Gu Z, Zhu Y, Jiang S, Xia G, Li C, Zhang X, Zhang J, Shen X. Tilapia head glycolipids reduce inflammation by regulating the gut microbiota in dextran sulphate sodium-induced colitis mice. Food Funct. 2020;11:3245-3255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 30. | Liu A, Lv H, Wang H, Yang H, Li Y, Qian J. Aging Increases the Severity of Colitis and the Related Changes to the Gut Barrier and Gut Microbiota in Humans and Mice. J Gerontol A Biol Sci Med Sci. 2020;75:1284-1292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 68] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 31. | Zhuang X, Tian Z, Li L, Zeng Z, Chen M, Xiong L. Fecal Microbiota Alterations Associated With Diarrhea-Predominant Irritable Bowel Syndrome. Front Microbiol. 2018;9:1600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 32. | Fung TC, Vuong HE, Luna CDG, Pronovost GN, Aleksandrova AA, Riley NG, Vavilina A, McGinn J, Rendon T, Forrest LR, Hsiao EY. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4:2064-2073. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 223] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 33. | Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, Niehus R, Staffas A, Dai A, Fontana E, Amoretti LA, Wright RJ, Morjaria S, Fenelus M, Pessin MS, Chao NJ, Lew M, Bohannon L, Bush A, Sung AD, Hohl TM, Perales MA, van den Brink MRM, Xavier JB. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588:303-307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 240] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 34. | Fuentes S, Rossen NG, van der Spek MJ, Hartman JH, Huuskonen L, Korpela K, Salojärvi J, Aalvink S, de Vos WM, D'Haens GR, Zoetendal EG, Ponsioen CY. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017;11:1877-1889. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 121] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 35. | Ma C, Wu W, Lin R, Ge Y, Zhang C, Sun S, Cong Y, Li X, Liu Z. Critical Role of CD6highCD4+ T Cells in Driving Th1/Th17 Cell Immune Responses and Mucosal Inflammation in IBD. J Crohns Colitis. 2019;13:510-524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, Owens BMJ, Uhlig HH, McMichael A; Oxford IBD Cohort Investigators; Bergthaler A, Teichmann SA, Keshav S, Powrie F. Circulating and Tissue-Resident CD4+ T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology 2017; 153: 1320-1337. e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 210] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 37. | Giuffrida P, Di Sabatino A. Targeting T cells in inflammatory bowel disease. Pharmacol Res. 2020;159:105040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 38. | Shi W, Zou R, Yang M, Mai L, Ren J, Wen J, Liu Z, Lai R. Analysis of Genes Involved in Ulcerative Colitis Activity and Tumorigenesis Through Systematic Mining of Gene Co-expression Networks. Front Physiol. 2019;10:662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 39. | Zhang XJ, Yuan ZW, Qu C, Yu XT, Huang T, Chen PV, Su ZR, Dou YX, Wu JZ, Zeng HF, Xie Y, Chen JN. Palmatine ameliorated murine colitis by suppressing tryptophan metabolism and regulating gut microbiota. Pharmacol Res. 2018;137:34-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 40. | Acovic A, Gazdic M, Jovicic N, Harrell CR, Fellabaum C, Arsenijevic N, Volarevic V. Role of indoleamine 2,3-dioxygenase in pathology of the gastrointestinal tract. Therap Adv Gastroenterol. 2018;11:1756284818815334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 41. | Shon WJ, Lee YK, Shin JH, Choi EY, Shin DM. Severity of DSS-induced colitis is reduced in Ido1-deficient mice with down-regulation of TLR-MyD88-NF-kB transcriptional networks. Sci Rep. 2015;5:17305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 42. | Shin JH, Lee YK, Shon WJ, Kim B, Jeon CO, Cho JY, Morse HC 3rd, Choi EY, Shin DM. Gut microorganisms and their metabolites modulate the severity of acute colitis in a tryptophan metabolism-dependent manner. Eur J Nutr. 2020;59:3591-3601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 43. | Kolls JK, McCray PB Jr, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol. 2008;8:829-835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 256] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 44. | Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 931] [Cited by in F6Publishing: 936] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 45. | Jones GR, Bain CC, Fenton TM, Kelly A, Brown SL, Ivens AC, Travis MA, Cook PC, MacDonald AS. Dynamics of Colon Monocyte and Macrophage Activation During Colitis. Front Immunol. 2018;9:2764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 46. | Thomson CA, McColl A, Graham GJ, Cavanagh J. Sustained exposure to systemic endotoxin triggers chemokine induction in the brain followed by a rapid influx of leukocytes. J Neuroinflammation. 2020;17:94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 47. | Sato N, Garcia-Castillo V, Yuzawa M, Islam MA, Albarracin L, Tomokiyo M, Ikeda-Ohtsubo W, Garcia-Cancino A, Takahashi H, Villena J, Kitazawa H. Immunobiotic Lactobacillus jensenii TL2937 Alleviates Dextran Sodium Sulfate-Induced Colitis by Differentially Modulating the Transcriptomic Response of Intestinal Epithelial Cells. Front Immunol. 2020;11:2174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |