Published online Nov 7, 2020. doi: 10.3748/wjg.v26.i41.6414
Peer-review started: July 20, 2020
First decision: August 8, 2020
Revised: August 17, 2020
Accepted: September 10, 2020
Article in press: September 10, 2020
Published online: November 7, 2020
Gastric cancer (GC) ranks as the third leading cause of cancer-related death worldwide. Epigenetic alterations contribute to tumor heterogeneity in early stages.
To identify the specific deoxyribonucleic acid (DNA) methylation sites that influence the prognosis of GC patients and explore the prognostic value of a model based on subtypes of DNA methylation.
Patients were randomly classified into training and test sets. Prognostic DNA methylation sites were identified by integrating DNA methylation profiles and clinical data from The Cancer Genome Atlas GC cohort. In the training set, unsupervised consensus clustering was performed to identify distinct subgroups based on methylation status. A risk score model was built based on Kaplan-Meier, least absolute shrinkage and selector operation, and multivariate Cox regression analyses. A test set was used to validate this model.
Three subgroups based on DNA methylation profiles in the training set were identified using 1061 methylation sites that were significantly associated with survival. These methylation subtypes reflected differences in T, N, and M category, age, stage, and prognosis. Forty-one methylation sites were screened as specific hyper- or hypomethylation sites for each specific subgroup. Enrichment analysis revealed that they were mainly involved in pathways related to carcinogenesis, tumor growth, and progression. Finally, two methylation sites were chosen to generate a prognostic model. The high-risk group showed a markedly poor prognosis compared to the low-risk group in both the training [hazard ratio (HR) = 2.24, 95% confidence interval (CI): 1.28-3.92, P < 0.001] and test (HR = 2.12, 95%CI: 1.19-3.78, P = 0.002) datasets.
DNA methylation-based classification reflects the epigenetic heterogeneity of GC and may contribute to predicting prognosis and offer novel insights for individualized treatment of patients with GC.
Core Tip: To address the epigenetic heterogeneity of gastric cancer, three subgroups based on deoxyribonucleic acid (DNA) methylation were identified and each subtype was associated with distinct survival and clinical features. A signature based on molecular subtypes of DNA methylation was built to predict the survival of gastric cancer patients, and showed good performance. This work may improve our understanding of the epigenetic landscape of gastric cancer and facilitate precision medicine for these patients.
- Citation: Bian J, Long JY, Yang X, Yang XB, Xu YY, Lu X, Sang XT, Zhao HT. Signature based on molecular subtypes of deoxyribonucleic acid methylation predicts overall survival in gastric cancer. World J Gastroenterol 2020; 26(41): 6414-6430
- URL: https://www.wjgnet.com/1007-9327/full/v26/i41/6414.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i41.6414
Gastric cancer (GC) ranks as the third leading cause of cancer-related deaths and is the fifth most commonly diagnosed cancer worldwide[1,2]. While curative resection, adjuvant or neoadjuvant therapy che-motherapy, and targeted therapies such as trastuzumab or ramucirumab may be curative treatment options for a select population of GC patients, high postoperative recurrence and metastasis make long-term survival dismal[3,4]. Studies have indicated that patients with metastasis had a survival of only 4 to 12 mo when treated with only best supportive care or chemotherapy[5]. Since GC is a genetically and epigenetically heterogeneous disease, identifying robust biomarkers is critical for early detection and survival prognosis. Conventional biomarkers, including carcinoembryonic antigen, carbohydrate antigen 19-9, carbohydrate antigen 72-4, and human epidermal growth factor receptor 2, have been widely used in clinical practice. Novel biomarkers, such as fibroblast growth factor receptor 2, vascular endothelial growth factor, E-cadherin, and microsatellite instability, have also been explored and shown to be valuable biomarkers[6,7]. However, due to inefficient specificity and sensitivity, limited novel biomarkers have been put into routine clinical practice. Therefore, it is needed to explore more efficient biomarkers based on genetic and epigenetic alterations. Deoxyribonucleic acid (DNA) methylation is a major epigenetic event that regulates gene transcription and maintains genome stability[8,9]. Oncogene hypomethylation and tumor suppressor gene hypermethylation are common methylation aberrations that have been shown to play important roles in cancer development, including the tumorigenesis of GC[10,11]. Detecting DNA methylation patterns and understanding the roles of these methylation events might help elucidate the underlying molecular mechanisms and pathogenesis of GC. Although there are abundant studies on the relationship between dysregulated DNA methylation and the prognosis of GC patients[12-14], individualized prognostic models based on a DNA methylation signature are lacking. In this study, we explored molecular subgroups of GC by integrating methylation and mRNA expression profile data, and generated a prognostic model comprising two DNA methylation sites. Our study may deepen our understanding and improve individualized therapies for GC.
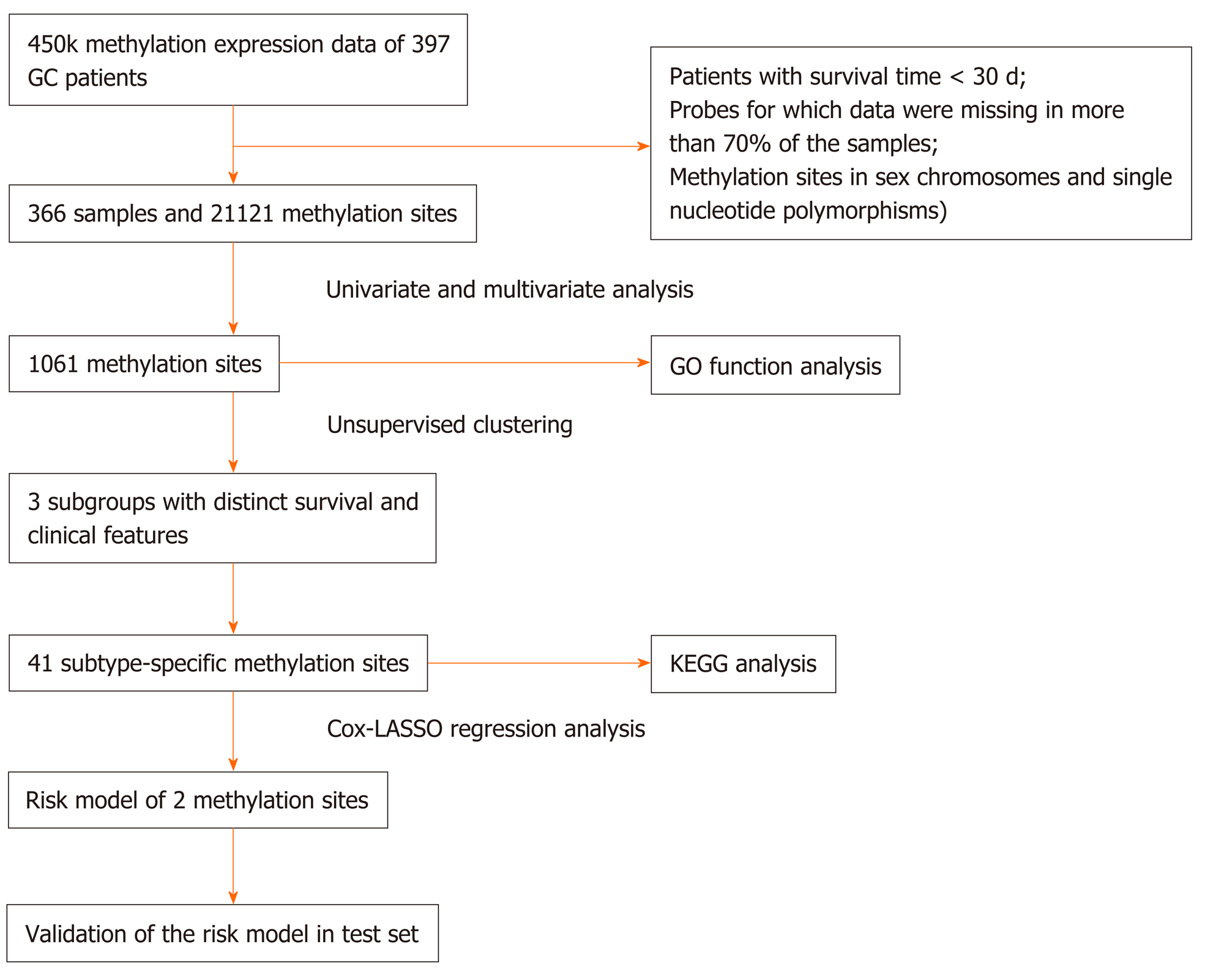
A total of 407 RNA-sequencing profiles (375 GC samples and 32 nontumor samples) and the corresponding clinical information were downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/, up to October 1, 2019, Supplementary Tables 1 and 2). We obtained DNA methylation profiles from the University of California Santa Cruz Cancer Browser (https://xena.ucsc.edu/), including the analysis of 397 patients with Illumina Infinium Human Methylation 450 platform. Methylation levels were quantified using beta values ranging from 0 to 1 (unmethylated to totally methylated). Samples with a follow-up time of less than 30 d or with a lack of clinical survival information were excluded. Probes for which CpG data were missing in more than 70% of the samples were removed. The K-nearest neighbors imputation procedure was used to impute the remaining probes with data not available. The ComBat algorithm in the sva R package[15] was used to remove batch effects by integrating all DNA methylation array data and incorporating batch and patient clinical information. Data with unstable methylation sites (CpGs in sex chromosomes and single nucleotide polymorphisms) were removed from the dataset. CpGs in promoter regions were selected and studied because the DNA methylation level in promoter regions are associated with gene expression. Promoter regions are located 2 kb upstream to 0.5 kb downstream from transcription start sites of genes. We selected samples for which RNA-sequencing data and DNA methylation data were available. In total, 366 samples and 21121 methylation sites were included in subsequent analyses. Moreover, 366 samples were then randomly stratified into the training set (n = 183) and test set (n = 183). DNA methylation-based subgroup analysis was performed in the training set and a risk score model was built, which was subsequently validated in the test set. The study flow chart is shown in Figure 1.
To determine GC molecular subtypes, we first selected CpG sites that were significantly associated with prognosis as classification features. Univariate and multivariate analyses were conducted using the Cox proportional hazard regression model. Univariate Cox proportional risk regression models were constructed for each CpG site, age, sex, T category, N category, M category, TNM stage, and survival time using methylation levels. The significant CpG sites obtained from univariate Cox proportional risk regression models were then analyzed using multivariate Cox proportional risk regression models. Consequently, N category, TNM stage, age, and sex, which were significant in the univariate survival analysis, were used as covariates in the multivariate analysis. CpG sites that were significant in both univariate and multivariate Cox regression analyses were selected as characteristic CpG sites. Univariate and multivariate analyses were performed with a P value of 0.05 as the cutoff.
Unsupervised consensus clustering using the ConsensusClusterPlus package in R[16] was performed to identify GC subgroups based on the characteristic CpG sites that were significant in both univariate and multivariate Cox regression analyses. To achieve higher intracluster similarity and lower intercluster similarity, we chose the k-means clustering algorithm with the Euclidean distance and a subsampling ratio of 0.8 for 100 iterations. The values of k where the magnitude of the relative change in area under the cumulative distribution function that began to fall were chosen as the optimal cluster numbers. The pheatmap package in R was used to generate the heatmap corresponding to the consensus clustering.
Differential analysis was conducted on the screened methylation profiles of each subtype to identify the specific methylation sites. A total of 1061 methylation sites among each subtype were analyzed. Every methylation site in each molecular subtype was compared with that in the other subtypes, and all methylation sites were analyzed using the Wilcoxon rank-sum test (false discovery rate < 0.05 and|log2 (fold change [FC])| > 1). Furthermore, the differential frequency of every CpG site in each subtype was further detected for the final screening of the CpG sites. One methylation site was defined as a specific methylation site if it satisfied the differential condition in only one subtype. The obtained specific methylation sites were subsequently subjected to genome annotations to identify their corresponding genes.
The overall survival (OS) for each DNA methylation subtype among GC patients was evaluated by Kaplan–Meier (K-M) analysis. The significance of differences among the clusters was assessed by the log-rank test. Associations between both the clinical and biological characteristics and DNA methylation clustering were analyzed using the chi-square test. Survival analyses were performed using the survival package in R. The statistical significance levels were all two-sided at P < 0.05, and the hazard ratio (HR) and 95% confidence interval (CI) were also calculated.
Corresponding genes in the promoter regions of these specific methylation sites were subjected to gene ontology (GO) and Kyoto encyclopedia of genes and genomes pathway enrichment analyses with the help of the clusterProfiler package in R[17]. Enriched functional annotations with an adjusted P value < 0.05 were considered significant.
Least absolute shrinkage and selection operator (LASSO) and multivariate Cox regression analyses were utilized to evaluate relationships between the specifically expressed methylation sites in each subtype and prognosis and to generate a prognostic prediction model for the training set. Using coefficients from multivariate Cox regression analysis as the weights, a prognostic prediction model was constructed through a linear combination of expression profile data of independent specific CpG methylation sites. The formula is as follows: Risk score = -1.483954476 × cg17398595 - 2.34637809416689 × cg20496643. Based on the risk score prediction model, GC patients were classified into low and high-risk groups with the optimal risk score as the cutoff value. X-tile[18] software was employed to determine the optimal cutoff value. The threshold for the risk score that was the output from the prediction model, which was utilized for separating patients into high and low-risk groups, was defined as the risk score that generated the largest value of χ² in the Mantel-Cox test. K-M and log-rank methods were used to evaluate the survival differences between high and low-risk patients. Time-dependent receiver operating characteristic curves were employed to measure the predictive performance, and the prognostic model was validated in the test set.
As described in the Materials and Methods, 21121 methylation sites were identified, of which 1507 CpG sites were identified as potential DNA methylation biomarkers for OS in GC patients using univariate Cox regression analysis (Supplementary Table 3). Univariate Cox proportional-hazards regression analysis revealed that N category (regional lymph nodes), TNM stage, age, and sex were significantly associated with OS (respective log-rank P values: 0.021503, 0.015607, 0.005479, and 0.033011). Subsequently, 1061 independent prognosis-associated CpG sites were obtained using multivariate Cox regression analysis of the 1507 methylation sites, with N category, TNM stage, age, and sex as covariates (Supplementary Table 4). These 1061 sites were significant in both univariate and multivariate analyses, and were selected as potential prognostic methylation sites.
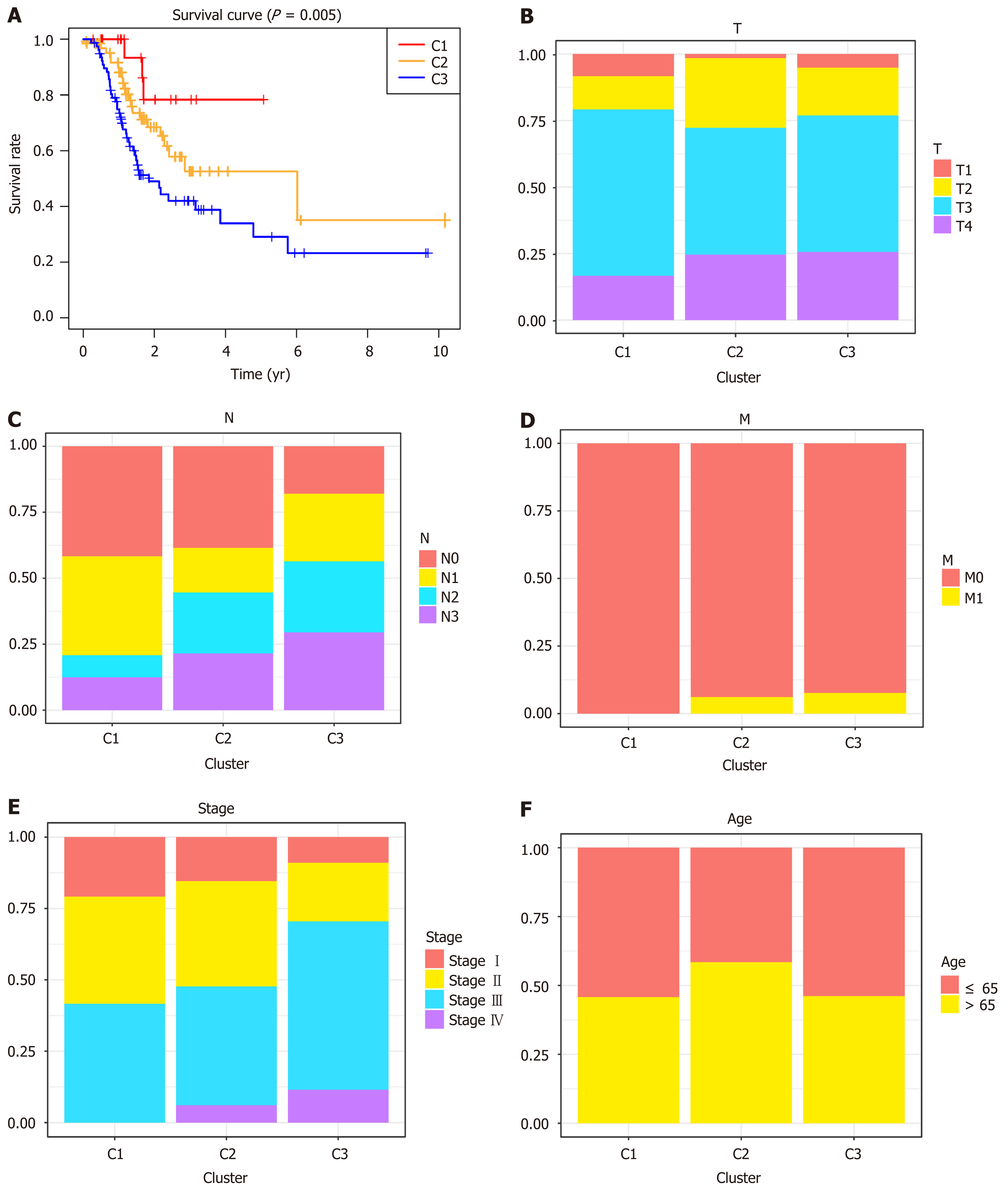
Unsupervised clustering of 1061 significant methylation sites was conducted to identify the molecular subtypes for subgroup classification in the training set. We then calculated the average cluster consensus and the coefficient of variation among clusters for each category number. The values of k where the largest magnitude of the relative change in area under the cumulative distribution function began to fall were chosen as the cluster numbers. After comprehensive consideration, k = 3 was selected to obtain three molecular subtypes for further analysis (Figure 2A). A heatmap of 1061 DNA methylation sites in three clusters was then constructed, with the T category, N category, M category, TNM stage, age, and DNA methylation subgroup as the annotations (Figure 2B). As shown in Figure 1B, although the abundance of most CpG sites was relatively low in each sample, there were obvious differences in the DNA methylation status among the three clusters. As shown in the boxplot, cluster 1 had the highest methylation level, while cluster 3 had the lowest methylation level (Supplementary Figure 1). K-M survival analysis showed significant differences in prognosis among the three clusters defined by DNA methylation unsupervised clustering (P = 0.005, Figure 3A). Cluster 1 had the best prognoses, while cluster 3 had the worst prognoses, indicating an association of lower methylation level with poorer survival for GC patients. To explore the clinical features of different methylation subtypes, we analyzed the distribution of T category, N category, M category, TNM stage, and age for the three clusters (Figure 3B-F). Compared to clusters 1 and 2, cluster 3 was prone to lymphatic invasion and metastasis and associated with a more advanced stage, which suggested an important role of neoadjuvant therapy for these patients. Notably, cluster 2 was associated with the lowest rate of T1 and high relevance with N3-4, indicating a more radical surgical approach in clinical practice. There were no differences observed in the grade or age among the three subtypes of GC patients.
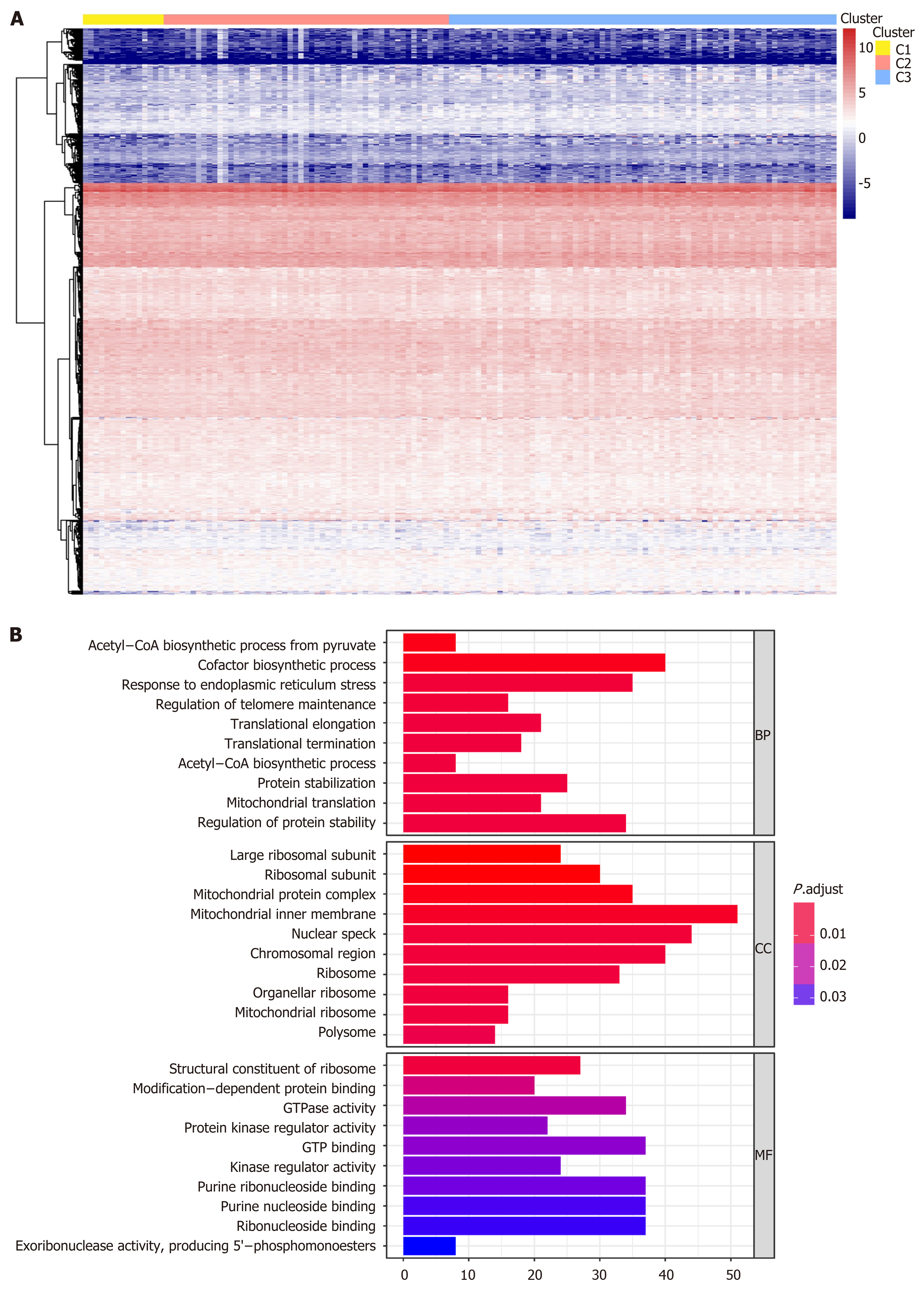
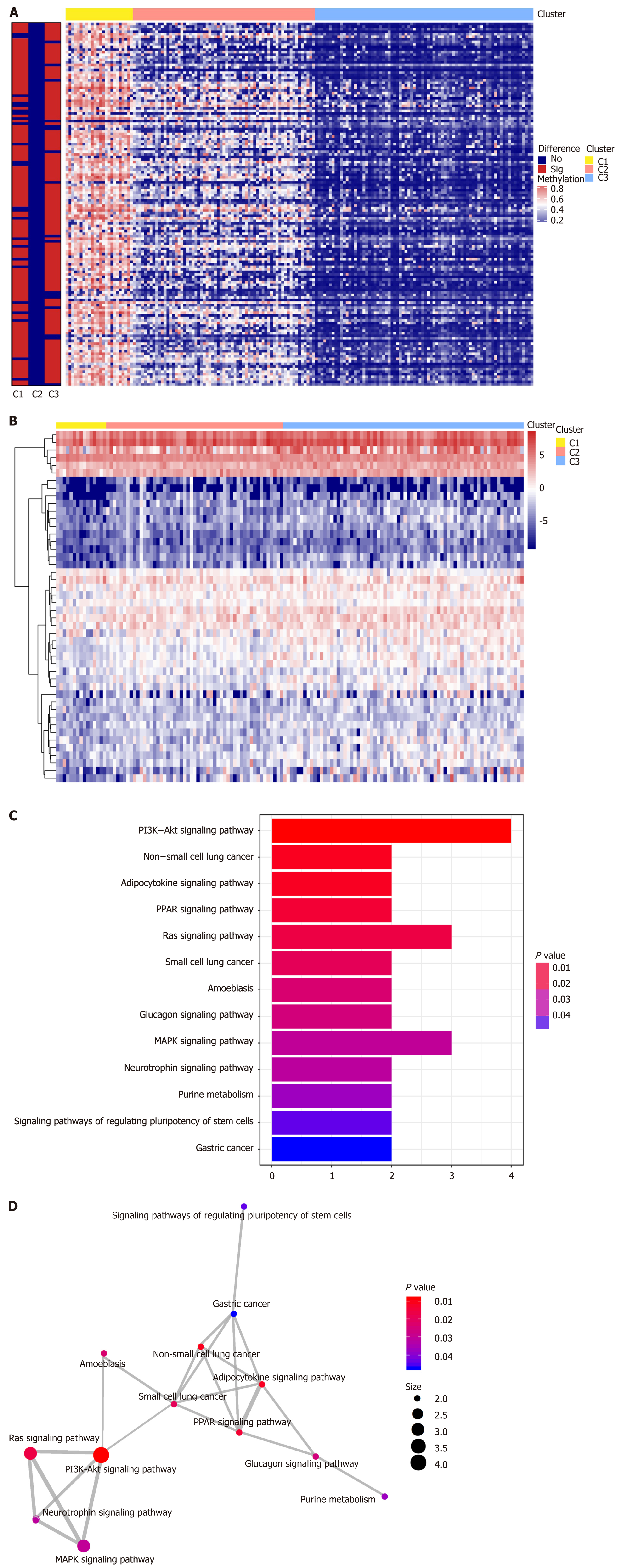
We performed genome annotations for the 1061 CpG sites described above and identified 1394 corresponding genes. The expression levels of these corresponding genes were visualized in a heatmap (Figure 4A). GO analyses were conducted to elucidate the functional characteristics of these promoter genes (P < 0.05, Figure 4B, Supplementary Table 5). GO functions of these genes were significantly enriched in protein synthesis and energy metabolism categories, such as “acetyl−CoA biosynthetic process from pyruvate”, “large ribosomal subunit”, and “structural constituent of ribosome”. The differences in the 1061 methylation sites in each subtype of GC were further analyzed using the Wilcoxon rank-sum test (false discovery rate < 0.05 and |log2 (fold change [FC]) > 1), and heatmap is presented in Figure 5A (Supplementary Table 6). We subsequently identified 41 subtype-specific CpG sites that were specifically hypermethylated or hypomethylated in only one subgroup (Supplementary Table 7). These 41 specific methylation sites were subsequently subjected to gene annotations, identifying 52 corresponding genes. To illustrate the expression of these specific methylation corresponding genes in the subgroups, the expression values of 167 samples in the training set for 46 of the 52 genes were obtained (Figure 5B). Distinct expression levels of these genes in specific subgroups were observed, indicating that the expression profiles of these specific methylation site-corresponding genes were consistent with the DNA methylation level. To gain a further understanding of the biological effects of the corresponding genes of these specific methylation sites, Kyoto encyclopedia of genes and genomes analysis was performed with a threshold of P < 0.05 (Figure 5C and D, Supplementary Table 8). As shown in Figure 5C, the top five signaling pathways are the PI3K-Akt signaling pathway, non-small cell lung cancer, adipocytokine signaling pathway, PPAR signaling pathway, and Ras signaling pathway. Crosstalk analysis showed close relationships among the 13 pathways. Most of these signaling pathways are reported to be involved in carcinogenesis and tumor growth and progression, indicating that the genes corresponding to the specific methylation sites are critical in the molecular mechanisms of GC development.
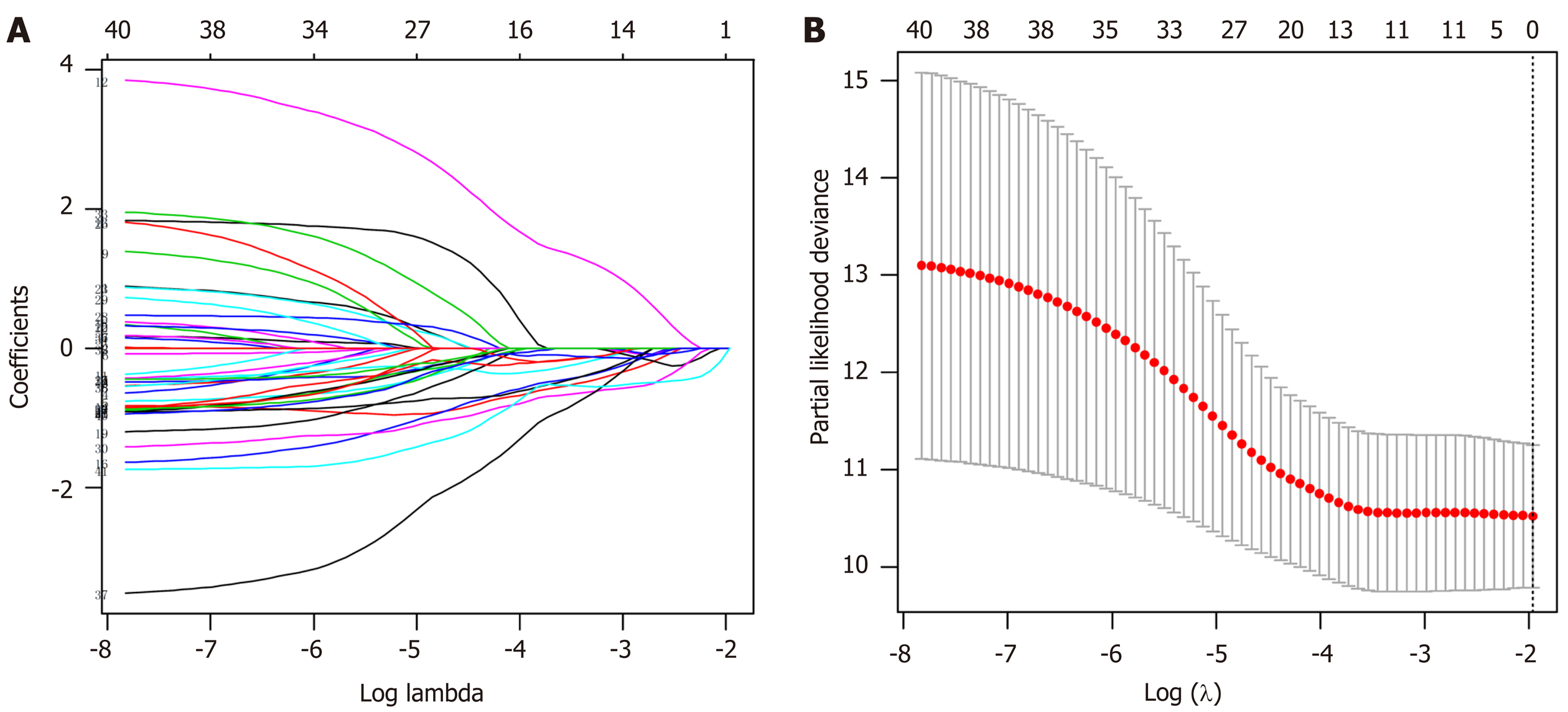
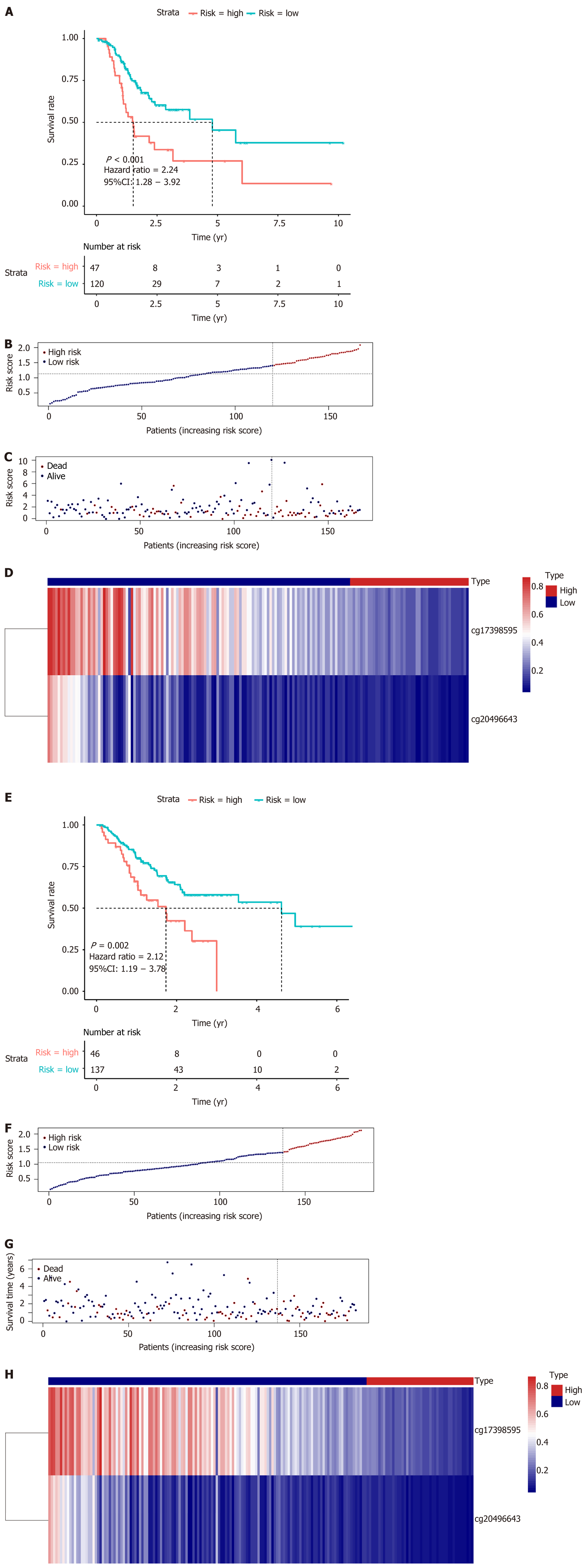
LASSO regression analysis is a penalized regression method that uses an L1 penalty to shrink regression coefficients toward zero, thereby eliminating a number of variables based on the principle that fewer predictors are selected when the penalty is larger[19]. Thus, seed methylation sites with nonzero coefficients were regarded as potential prognostic predictors. Based on 1000 iterations of Cox-LASSO regression analysis with 10-fold cross-validation using the glmnet package in R, the seed methylation sites were shrunk into multiple-site sets. Methylation sites with nonzero coefficients were considered potential prognostic genes. The 41 selected DNA methylation sites were analyzed by 1000 iterations of Cox-LASSO regression to reduce the number. Applying LASSO regression analysis, in which the selected DNA methylation sites were required to appear 500 times out of 1000 repetitions, five methylation sites were selected as prognostic CpGs (Figure 6A and B). Then, using the regression coefficient from a multivariate Cox proportional hazard model, we established a model including two methylation sites by Akaike Information Criterion in a stepwise algorithm. According to the optimal cutoff value, the patients were stratified into high and low-risk groups. High-risk patients showed significantly worse OS (HR = 2.24, 95%CI: 1.28-3.92, P < 0.001) than low-risk patients (Figure 7A). Figure 7B-D displays methylation levels of CpG sites and risk score distributions. Methylation levels for the two methylation sites significantly decreased as risk scores increased. Receiver operating characteristic analysis was performed to determine the specificity and sensitivity of the prognostic model. The time-dependent area under the curves for the 3-year OS rates for GC patients with the prognostic model were 0.610 (Supplementary Figure 2A). The predictive ability and stability of the prognostic model were further tested using 183 GC samples with OS time and survival status in the test set. The patients in the test set were classified into high and low-risk groups using the same formula and cutoff obtained from the training set. Consistent with the results in the training set, patients in the high-risk group in the testing set had a significantly shorter median OS than those in the low-risk group (HR = 2.12, 95%CI: 1.19-3.78, P = 0.002) (Figure 7E). Figure 7F-H shows the distribution of risk scores and CpG site methylation levels. The time-dependent area under the curve of the 3-year OS rate with the prognostic model for GC patients was 0.696 (Supplementary Figure 2B).
GC is one of the most common malignancies, causing one of the highest public health burdens[1,20]. Studies have shown that GC carcinogenesis is a multistep and multifactorial process caused by genetic changes and epigenetic alterations[2,21]. GC is characterized by accumulated genomic modifications, including somatic mutations and genomic amplifications and deletions[22]. However, evidence has shown that both genomic aberrations and Helicobacter pylori-induced precursors are associated with multiple epigenetic changes, such as hypermethylation of tumor suppressors and hypomethylation of oncogenes[12,21]. For instance, Helicobacter pylori can induce methylation of multiple CpG islands in GC patients, which subsequently increases genome instability by stimulating activation-induced cytidine deaminase or altering microRNA expression[23]. Therefore, it is important to identify key mechanisms involved in epigenetic alterations and elucidate the role of DNA methylation in GC development and progression.
Epigenetic changes, including DNA and histone modifications, can result in dysregulated expression of tumor suppressor genes and oncogenes. Aberrant methylation changes occur frequently in human cancers. For instance, the DNA methyltransferase family is responsible for DNA methylation, and altered expression of DNA methyltransferase has been shown to be involved in the pathogenesis of GC[13,24]. There is evidence that altered DNA methylation is an early event in the development and progression of GC[25], and these aberrant DNA methylations can be targeted by DNA methylation inhibitors[26]. Studies have shown that epigenetic changes occurred prior to genome alterations in normal and nonneoplastic gastric mucosa, and abnormal methylation levels were associated with an increased risk of GC[27-29]. Methylation of tumor suppressor genes, such as RUNX3, CDH1, APC, CHFR, DAPK, and GSTP1, is associated with the onset of GC and plays important roles in the early stages of tumor development. DNA methylation alterations have not only been associated with GC development in the early stage, but can also be useful for survival prognosis. For example, GC patients with the hypermethylation of MADGA2, which is a tumor suppressor, were associated with significantly decreased survival time[14]. Clarifying altered DNA methylation can aid in the early diagnosis and survival prognosis of GC. As in most cancers, GC is a heterogeneous disease with distinct phenotypes. Integrative molecular subtype analysis of cancer can provide insights into carcinogenesis, diagnosis, and prognosis. Recent studies have highlighted the predictive role of methylation patterns in different cancers[30-32]. However, the association between methylation status and survival prognosis is controversial in different studies. While some studies indicated that GC hypermethylation was associated with a good prognosis[33,34], others reported an association with poor survival[35,36]. A meta-analysis of 918 patients showed that hypermethylation of CpG islands was significantly associated with a poor 5-year survival; however, the results were less convincing due to great heterogeneity among the included studies[37].
Our study contributed to the understanding of the epigenetic landscape of GC. In this study, we identified three subtypes of GC based on DNA methylation, which were characteristic with distinct prognoses and clinical features. These molecular subtypes of GC may shed light on future clinical stratification and subtype-based targeted therapies. We focused on specific DNA methylation markers and analyzed DNA methylation prognosis subgroups of GC. We attempted to address the relations between specific methylation status and prognosis by developing a classification model that integrated two DNA methylation biomarkers for the prognostic evaluation of GC patients. Moreover, our signature is based on two specific methylation sites and is easy to test in clinical practice, with considerable cost-effectiveness. However, our research has limitations because it was retrospective, and our results need to be further confirmed by prospective studies. Moreover, due to the relatively small number of patients, the efficiency of the prognostic model should be further validated using a large number of GC patients.
In summary, our study identified three molecular subtypes based on DNA methylation in GC and established a prognostic prediction model with prognosis-specific methylation sites. These results may help improve outcome prediction, and facilitate precision therapy for patients with GC.
Gastric cancer (GC) is a heterogeneous disease with genetic and epigenetic alterations. Robust biomarkers for management and survival prognosis of GC patients are lacking. Deoxyribonucleic acid (DNA) methylation is a major epigenetic event that participates in early stage of GC and is suggested to be associated with survival in many cancers including GC.
Exploring molecular subtypes of GC can improve understanding of this heterogeneous cancer and contribute to better management and prognosis prediction. Studies on DNA methylation subtypes of GC are lacking.
To identify the specific DNA methylation sites that influence the prognosis of GC patients by integrating epigenetic and clinical information. We also aimed to establish a prognostic model based on subtypes of DNA methylation.
Data of GC patients were obtained from The Cancer Genome Atlas and the University of California Santa Cruz cancer browser. Prognostic DNA methylation sites were identified by integrating DNA methylation profiles and clinical data. We used unsupervised clustering to identify distinct subgroups based on methylation status. A risk score model was built and further validated in a test set.
In this study, we identified three subtypes based on DNA methylation profiles using methylation sites that were significantly associated with survival. These methylation subtypes were associated with clinical features, patient outcomes, and potential responses to therapy. Enrichment analysis of specific hyper- or hypomethylation sites revealed that they were mainly involved in pathways related to carcinogenesis and tumor growth and progression. A prognostic model consisting two methylation sites was subsequently generated. The high-risk group showed a significantly poorer prognosis compared to the low-risk group in both the training (hazard ratio = 2.24, 95% confidence interval: 1.28-3.92, P < 0.001) and test (hazard ratio = 2.12, 95% confidence interval: 1.19-3.78, P = 0.002) sets. More samples are needed to optimize the model performance.
This study indicates that DNA methylation-based classification reflects the epigenetic heterogeneity of GC. A prediction model based on methylation subtypes can predict the OS of GC patients.
Our study can help predict prognosis and increase our understanding of the heterogeneity of GC patients. This is a retrospective analysis of GC patients from public database, so prospective studies are needed to validated the findings.
We thank Yu Lin for assistance with the data interpretation.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Czubkowski P, Kositamongkol P, Lee S, Schievenbusch S S-Editor: Zhang L L-Editor: Wang TQ P-Editor: Zhang YL
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50873] [Article Influence: 8478.8] [Reference Citation Analysis (44)] |
| 2. | Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-2664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1282] [Cited by in F6Publishing: 1367] [Article Influence: 170.9] [Reference Citation Analysis (0)] |
| 3. | Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22:2403-2414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 310] [Cited by in F6Publishing: 361] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 4. | Ajani JA, D'Amico TA, Almhanna K, Bentrem DJ, Chao J, Das P, Denlinger CS, Fanta P, Farjah F, Fuchs CS, Gerdes H, Gibson M, Glasgow RE, Hayman JA, Hochwald S, Hofstetter WL, Ilson DH, Jaroszewski D, Johung KL, Keswani RN, Kleinberg LR, Korn WM, Leong S, Linn C, Lockhart AC, Ly QP, Mulcahy MF, Orringer MB, Perry KA, Poultsides GA, Scott WJ, Strong VE, Washington MK, Weksler B, Willett CG, Wright CD, Zelman D, McMillian N, Sundar H. Gastric Cancer, Version 3.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2016;14:1286-1312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 640] [Cited by in F6Publishing: 632] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 5. | Glimelius B, Ekström K, Hoffman K, Graf W, Sjödén PO, Haglund U, Svensson C, Enander LK, Linné T, Sellström H, Heuman R. Randomized comparison between chemotherapy plus best supportive care with best supportive care in advanced gastric cancer. Ann Oncol. 1997;8:163-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 592] [Cited by in F6Publishing: 586] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 6. | Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24:2818-2832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 230] [Cited by in F6Publishing: 260] [Article Influence: 43.3] [Reference Citation Analysis (5)] |
| 7. | Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, Kim J, Chatila W, Akbani R, Kanchi RS, Rabkin CS, Willis JE, Wang KK, McCall SJ, Mishra L, Ojesina AI, Bullman S, Pedamallu CS, Lazar AJ, Sakai R; Cancer Genome Atlas Research Network; Thorsson V; Bass AJ; Laird PW. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell 2018; 33: 721-735. e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 318] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 8. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2210] [Cited by in F6Publishing: 2104] [Article Influence: 105.2] [Reference Citation Analysis (0)] |
| 9. | Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1819] [Cited by in F6Publishing: 1897] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 10. | Huang KK, Ramnarayanan K, Zhu F, Srivastava S, Xu C, Tan ALK, Lee M, Tay S, Das K, Xing M, Fatehullah A, Alkaff SMF, Lim TKH, Lee J, Ho KY, Rozen SG, Teh BT, Barker N, Chia CK, Khor C, Ooi CJ, Fock KM, So J, Lim WC, Ling KL, Ang TL, Wong A, Rao J, Rajnakova A, Lim LG, Yap WM, Teh M, Yeoh KG, Tan P. Genomic and Epigenomic Profiling of High-Risk Intestinal Metaplasia Reveals Molecular Determinants of Progression to Gastric Cancer. Cancer Cell 2018; 33: 137-150. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 11. | Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics. 2008;9:394-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 173] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 12. | Ding WJ, Fang JY, Chen XY, Peng YS. The expression and clinical significance of DNA methyltransferase proteins in human gastric cancer. Dig Dis Sci. 2008;53:2083-2089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Yang J, Wei X, Wu Q, Xu Z, Gu D, Jin Y, Shen Y, Huang H, Fan H, Chen J. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol Med Rep. 2011;4:1139-1143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Wang K, Liang Q, Li X, Tsoi H, Zhang J, Wang H, Go MY, Chiu PW, Ng EK, Sung JJ, Yu J. MDGA2 is a novel tumour suppressor cooperating with DMAP1 in gastric cancer and is associated with disease outcome. Gut. 2016;65:1619-1631. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882-883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2446] [Cited by in F6Publishing: 3074] [Article Influence: 256.2] [Reference Citation Analysis (0)] |
| 16. | Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572-1573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1686] [Cited by in F6Publishing: 2923] [Article Influence: 208.8] [Reference Citation Analysis (0)] |
| 17. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11591] [Cited by in F6Publishing: 16524] [Article Influence: 1377.0] [Reference Citation Analysis (0)] |
| 18. | Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252-7259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1947] [Cited by in F6Publishing: 2611] [Article Influence: 137.4] [Reference Citation Analysis (0)] |
| 19. | Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 42] [Reference Citation Analysis (0)] |
| 20. | Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 900] [Cited by in F6Publishing: 1163] [Article Influence: 116.3] [Reference Citation Analysis (0)] |
| 21. | Fattahi S, Golpour M, Amjadi-Moheb F, Sharifi-Pasandi M, Khodadadi P, Pilehchian-Langroudi M, Ashrafi GH, Akhavan-Niaki H. DNA methyltransferases and gastric cancer: insight into targeted therapy. Epigenomics. 2018;10:1477-1497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, Ho SL, Chan AK, Man JL, Foglizzo V, Ng MK, Chan AS, Ching YP, Cheng GH, Xie T, Fernandez J, Li VS, Clevers H, Rejto PA, Mao M, Leung SY. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573-582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 694] [Cited by in F6Publishing: 765] [Article Influence: 76.5] [Reference Citation Analysis (0)] |
| 23. | Graham DY. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology 2015; 148: 719-31. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 287] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 24. | Cui H, Hu Y, Guo D, Zhang A, Gu Y, Zhang S, Zhao C, Gong P, Shen X, Li Y, Wu H, Wang L, Zhao Z, Fan H. DNA methyltransferase 3A isoform b contributes to repressing E-cadherin through cooperation of DNA methylation and H3K27/H3K9 methylation in EMT-related metastasis of gastric cancer. Oncogene. 2018;37:4358-4371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 25. | Watanabe Y, Kim HS, Castoro RJ, Chung W, Estecio MR, Kondo K, Guo Y, Ahmed SS, Toyota M, Itoh F, Suk KT, Cho MY, Shen L, Jelinek J, Issa JP. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology. 2009;136:2149-2158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 26. | Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 763] [Cited by in F6Publishing: 771] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 27. | Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, Sugimura T, Ichinose M, Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989-995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 460] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 28. | Tahara T, Shibata T, Nakamura M, Yamashita H, Yoshioka D, Okubo M, Yonemura J, Maeda Y, Maruyama N, Kamano T, Kamiya Y, Fujita H, Nakagawa Y, Nagasaka M, Iwata M, Hirata I, Arisawa T. Increased number of CpG island hypermethylation in tumor suppressor genes of non-neoplastic gastric mucosa correlates with higher risk of gastric cancer. Digestion. 2010;82:27-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Nakajima T, Maekita T, Oda I, Gotoda T, Yamamoto S, Umemura S, Ichinose M, Sugimura T, Ushijima T, Saito D. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317-2321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 130] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 30. | Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep 2018; 25: 1066-1080. e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 31. | Holm K, Staaf J, Lauss M, Aine M, Lindgren D, Bendahl PO, Vallon-Christersson J, Barkardottir RB, Höglund M, Borg Å, Jönsson G, Ringnér M. An integrated genomics analysis of epigenetic subtypes in human breast tumors links DNA methylation patterns to chromatin states in normal mammary cells. Breast Cancer Res. 2016;18:27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 32. | Long J, Chen P, Lin J, Bai Y, Yang X, Bian J, Lin Y, Wang D, Yang X, Zheng Y, Sang X, Zhao H. DNA methylation-driven genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Theranostics. 2019;9:7251-7267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 33. | An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, Ajani JA, Rashid A, Hamilton SR, Wu TT. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11:656-663. [PubMed] [Cited in This Article: ] |
| 34. | Shigeyasu K, Nagasaka T, Mori Y, Yokomichi N, Kawai T, Fuji T, Kimura K, Umeda Y, Kagawa S, Goel A, Fujiwara T. Clinical Significance of MLH1 Methylation and CpG Island Methylator Phenotype as Prognostic Markers in Patients with Gastric Cancer. PLoS One. 2015;10:e0130409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 35. | Chen HY, Zhu BH, Zhang CH, Yang DJ, Peng JJ, Chen JH, Liu FK, He YL. High CpG island methylator phenotype is associated with lymph node metastasis and prognosis in gastric cancer. Cancer Sci. 2012;103:73-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 36. | Park SY, Kook MC, Kim YW, Cho NY, Jung N, Kwon HJ, Kim TY, Kang GH. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch. 2010;457:415-422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Powell AGMT, Soul S, Christian A, Lewis WG. Meta-analysis of the prognostic value of CpG island methylator phenotype in gastric cancer. Br J Surg. 2018;105:e61-e68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |