Published online Sep 21, 2020. doi: 10.3748/wjg.v26.i35.5248
Peer-review started: May 28, 2020
First decision: June 18, 2020
Revised: June 30, 2020
Accepted: August 26, 2020
Article in press: August 26, 2020
Published online: September 21, 2020
Processing time: 111 Days and 12.3 Hours
Lesions missed by colonoscopy are one of the main reasons for post-colonoscopy colorectal cancer, which is usually associated with a worse prognosis. Because the adenoma miss rate could be as high as 26%, it has been noted that endoscopists with higher adenoma detection rates are usually associated with lower adenoma miss rates. Artificial intelligence (AI), particularly the deep learning model, is a promising innovation in colonoscopy. Recent studies have shown that AI is not only accurate in colorectal polyp detection but can also reduce the miss rate. Nevertheless, the application of AI in real-time detection has been hindered by heterogeneity of the AI models and study design as well as a lack of long-term outcomes. Herein, we discussed the principle of various AI models and systematically reviewed the current data on the use of AI on colorectal polyp detection and miss rates. The limitations and future prospects of AI on colorectal polyp detection are also discussed.
Core Tip: This review highlights the results of recent studies on the use of artificial intelligence for the detection of colorectal polyps and its role in reducing missed lesions during colonoscopy.
- Citation: Lui TKL, Leung WK. Is artificial intelligence the final answer to missed polyps in colonoscopy? World J Gastroenterol 2020; 26(35): 5248-5255
- URL: https://www.wjgnet.com/1007-9327/full/v26/i35/5248.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i35.5248
Colorectal cancer (CRC) is the third most common cancer worldwide. In 2015, there were 1.7 million new cases, resulting in more than 800000 deaths worldwide[1]. Screening colonoscopy and polypectomy have been shown to be effective in reducing the incidence of colorectal cancer as well as the associated cancer mortalities[2,3]. However, colonoscopy is not risk proof, and CRC can still develop within a short interval after a negative colonoscopy for cancer. In particular, post-colonoscopy colorectal cancer (PCCRC) is the preferred term used to define cancers appearing after a colonoscopy in which no cancer is diagnosed. Specifically, PCCRC can be further subdivided into “interval cancer,” where cancer is identified before the next recommended screening or surveillance examination. PCCRC, or interval cancer, could account for up to 9% of all colorectal cancers and is usually associated with an adverse outcome[4]. Recent studies showed that missed polyps and adenoma by colonoscopy accounted for at least 50% of all PCCRCs[5-7]. Therefore, ways to minimize missed lesions during colonoscopy are of utmost importance to maintain the quality and effectiveness of colonoscopy in preventing CRC.
It was shown in a recent meta-analysis that up to 26% of colonoscopies could have missed adenomas[8]. While many factors could affect the adenoma miss rate (AMR), the endoscopist factor was recognized to be one of the main determinants of AMR. High adenoma detection rate (ADR), high adenomas per index colonoscopy and high adenomas per positive index colonoscopy of the endoscopist were all shown to be negatively associated with AMR[8]. In particular, higher adenomas per positive index colonoscopy was independently associated with a lower advanced adenoma miss rate, which was an important predictor for PCCRC. Intuitively, ways to improve the ADR could also help to minimize AMR[9].
While “to err is human,” the mitigation of human factors, such as distraction, fatigue, impaired level of alertness, visual perception and recognition errors, may be the key to improving adenoma detection and hence reducing miss rates[10-14]. Additionally, patient factors, mainly poor bowel preparation, were also associated with lower ADR[15] and higher AMR[8]. However, there was minimal difference between fair- and good-quality bowel preparation in ADR and AMR[8], implying that at least fair bowel preparation should be achieved. Adequate withdrawal time, a minimum of 6 min, is another important quality measure to optimize ADR and AMR[16-18]. Another factor that would improve the ADR and reduce the AMR was the use of auxiliary techniques. There are a large number of auxiliary techniques, including second-pass colonoscopy[19], retroflexion in right-sided colon[20], water-aided colonoscopy[21], team detection approach (endoscopist and experienced nurse)[13,22], wide-angle endoscopy[23], high-definition endoscopy with or without a special imaging technique[24-26] and add-on devices[27], that have been reported to increase the ADR.
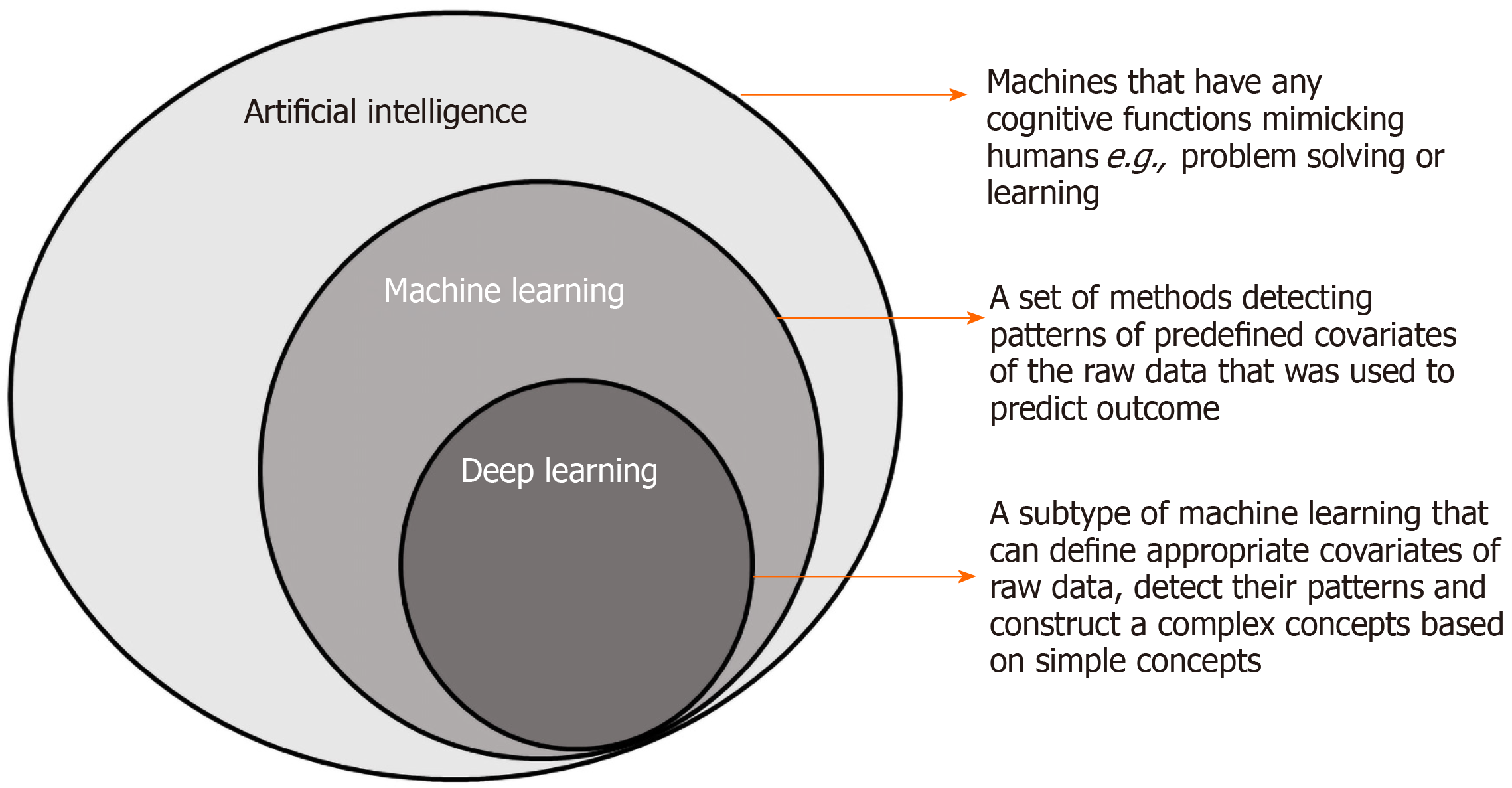
Artificial intelligence (AI) has been applied in the medical field since the early 1950s. AI is defined as any machine that has cognitive functions mimicking humans, e.g., problem solving or learning[28]. The machine learning model, which is a subtype of AI, is characterized by a set of methods that can automatically detect patterns in data and then use the uncovered patterns to predict outcomes[29]. Conventional AI systems utilize a supervised type of machine learning model that extracts the covariates of training data to achieve pattern recognition or classification. It is important to note that each piece of information included in the representation of the patient is known as a covariate, and the traditional type of machine learning, e.g., logistic regression, only examines the relationships of “predefined covariates” with the outcome[30]. Nevertheless, the machine learning model cannot change the way in which covariates are defined. The deep learning model actually solves this problem by defining covariates and builds up complex concepts from simple covariates, which is particularly useful in image classification and object location because features of a group of similar subjects can be complex and difficult to be defined by humans[30] (Figure 1).
In recent years, the deep learning model was increasingly used in the detection and localization of colorectal polyps. Once training data were provided with proper labels, the deep learning model could automatically extract the important features in the training data for differentiation and classification. Without the need of human intervention or indication, the internal parameters of each “neuron” in a single layer would be tuned towards a model with the least degree of error[31]. The most common architecture used in the deep learning model of early colonoscopy studies was convolutional neural networks, which mimics the structure of the human brain and contains multiple layers with “artificial neurons” under each layer. The convoluted layers actually act as a filter for extraction of the important features from the original image or data. The pooling layers can downsize the parameters of the layers to streamline the underlying computation. Finally, with the fully connected layers, these features are combined together to create a model to classify different outputs[32,33].
Our meta-analysis of recently published AI studies on colorectal polyp detection suggested that a well-designed AI system could achieve more than 90% accuracy[34-36]. Compared to a traditional machine learning based algorithm, studies using the deep learning model were found to have high accuracy (up to 91%) with a pooled sensitivity of 94% and a specificity of 92% on the detection of colorectal polyps[36].
As yet, most of the previously published studies have been retrospective in nature, and there have been limited high-quality prospective real-time studies on the use of AI in actual patients until recently. The first randomized controlled trial was reported in 2019 by Wang et al[37]. They showed that the use of real-time automatic polyp detection system (CADe) based on a deep learning architecture can increase the ADR in patients with a low prevalence of adenoma (20%-30%). Among the 1130 patients randomized, the ADR of the CADe group was significantly higher than that of the conventional colonoscopy group (0.29 vs 0.20, P < 0.001). The mean numbers of polyp and adenoma detected in the CADe group also increased from 0.50 to 0.95 (P < 0.001) and from 0.31 to 0.53 (P < 0.001), respectively, when compared with conventional colonoscopy.
Five recently published randomized controlled trials (RCTs) in 2020 again confirmed that AI-assisted colonoscopy significantly increased the adenoma detection rate when compared to conventional colonoscopy. Wang et al[38] further reported another RCT to compare a CADe system with a sham system. Again, the CADe system had a significantly higher ADR than the sham system (34% vs 28%, P = 0.03). In the same trial, adenoma or sessile serrated adenoma missed by endoscopists were characterized by isochromatic color, flat shape and located at the edge of the visual field or even partly behind colonic folds. Another randomized controlled trial by Repici et al[39] involving three centers in Italy also found that the CADe system was associated with a higher ADR with an odds ratio (OR) of 1.30 (95%CI: 1.14-1.45). Subgroup analysis showed that the performance of the CADe system was not affected by the size, shape and location of the polyps.
In addition to polyp location systems, Gong et al[40] reported a CADe system that aimed to monitor real-time withdrawal speed and to minimize blind spots during withdrawal. Their study showed that the ADR also improved from 8% to 16% (P = 0.001) with the CADe system. Similarly, Su et al[41] reported an automatic quality control system on colorectal polyp and adenoma detection that would also remind the endoscopist of the withdrawal time and speed and the need to re-examine unclear colonic segments on top of a polyp localization system. The system was found to have a significantly higher ADR than conventional colonoscopy (28.9% vs 16.5%, P < 0.001).
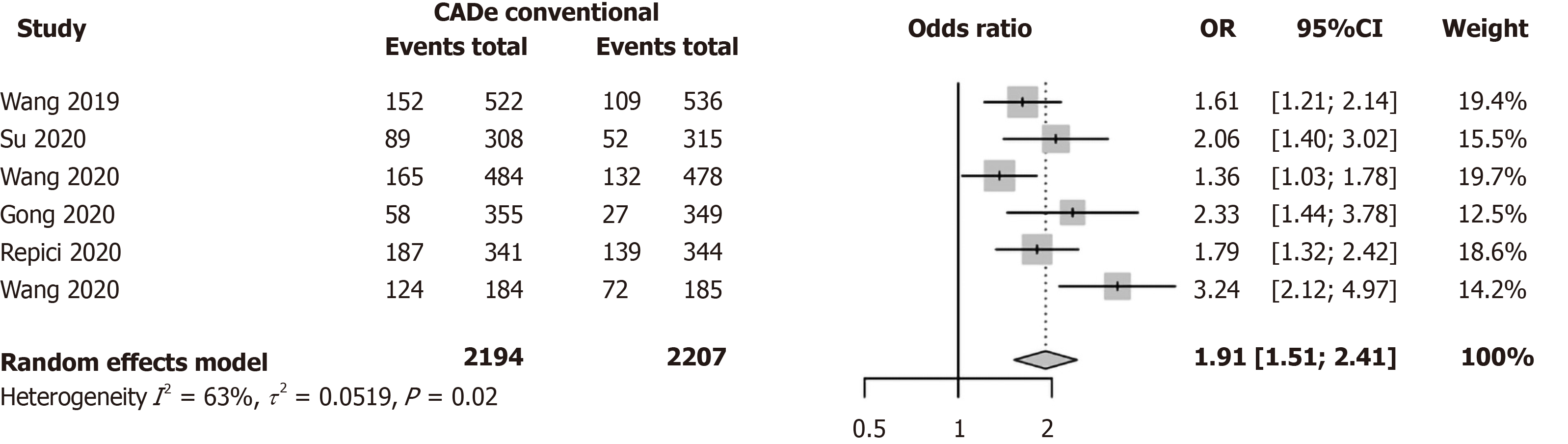
In view of these newly available RCTs after the publication of our meta-analysis[36], we have summarized the results of the latest prospective RCTs here in a new meta-analysis. In this meta-analysis of six RCTs, the pooled OR for the improvement of ADR was 1.91 (95%CI: 1.51-2.41) under a random effects model with a heterogeneity of I2 = 63% (Figure 2). Hence, there is convincing data from RCTs to show that the existing AI models could already help to boost the ADR by 90%.
In addition to its role in enhanced colorectal polyp detection, there are emerging data to suggest that AI could also help to reduce missed lesions during colonoscopy. In our recent study[42], we showed that the validated real-time deep learning AI model could help endoscopists to prevent missed colorectal lesions. We first applied the validated AI system to review 65 videos of tandem examinations of the proximal colon (from cecum to splenic flexure) and found that the AI system could detect up to 79.1% of adenomas that were missed by the endoscopist during the first-pass examination. In the second part of the prospective study, the same deep learning AI model was able to detect missed adenomas in 26.9% of patients during real-time examination. In multivariable analysis, missed adenomas were associated with findings of multiple polyps during colonoscopy (adjusted OR, 1.05) or colonoscopy performed by less-experienced endoscopists (adjusted OR, 1.30).
A recent single-center RCT by Wang et al[43] also showed that the use of CADe-assisted colonoscopy can reduce AMR from 40.0% to 14.0%. In particular, there were significant improvements in the ascending, transverse and descending colon. However, the AMR in this RCT (up to 40%) was much higher than previously reported. Therefore, a multicenter trial would still be required to validate this finding.
While supporting the role of AI in reducing missed lesions, these results suggested that the main reason for missed adenoma could still be due to human factors, as nearly 80% of these missed lesions were actually shown on screen and were not picked up by endoscopists for various reasons, such as inexperience, fatigue or distraction. Therefore, the AI could serve as an additional “eye” for the endoscopist with which distraction and fatigue would never occur.
However, our study also showed that approximately 20% of missed adenomas were still not detected even by AI. These missed lesions were usually not shown “on screen.” They were located behind a fold or at a difficult flexure position or hidden under the fecal contents in patients with poor bowel preparation. In a recent meta-analysis by Zhao et al[8] including 43 studies and 15000 tandem colonoscopies, the use of auxiliary techniques and good bowel preparation were associated with fewer missed adenomas. Intuitively, the combination of AI and auxiliary devices in the presence of satisfactory bowel preparation may be necessary to completely eliminate the risk of missed colonic lesions during colonoscopy.
In addition to detection of colorectal polyps, AI has also been shown to be accurate in histology prediction and polyp characterization in a number of studies[36]. Although there was a high degree of heterogeneity in the algorithms and design, along with potential selection biases, studies using a deep learning model as a backbone generally performed better than those using other types of algorithms. A study by Byrne et al[44] showed that the use of a deep learning model can achieve a 94% accuracy in the real-time classification of polyps. A similar result was reproduced by a study[45] using magnifying colonoscopy, and both studies used narrow-band imaging as the imaging technique. Our recent meta-analysis further showed that the pooled accuracy from studies using narrow-band imaging was generally better than that of non-narrow-band imaging studies in histology characterization[36].
Although there have been promising prospective trials supporting the use of AI in real-time polyp detection during colonoscopy, there are a number of issues to be addressed before AI can be implemented in routine clinical practice. Because the algorithms of AI and deep learning models are still evolving and there is substantial heterogeneity among different models and training data[36], an independent prospective validation would be required for each AI system. The latest guideline issued by the European Society of Gastrointestinal Endoscopy suggested that the possible incorporation of computer-aided diagnosis (detection and characterization of lesions) into colonoscopy should be supported by an acceptable and reproducible accuracy for colorectal neoplasia, as demonstrated in high-quality multicenter clinical studies[14]. Another important question regarding the use of AI in colonoscopy is the actual impact on long-term clinical outcomes. It is still unknown whether the use of AI-assisted colonoscopy can decrease the PCCRC rate or lengthen the current recommended surveillance interval after colonoscopy, which would require long-term prospective cohort studies to address.
The current role of AI in colonoscopy is possibly to act as a virtual assistant to the endoscopist during real-time colonoscopy, particularly in withdrawal time monitoring and polyp detection. The prospect of a fully automated independent colonoscopy system is still too premature at this stage. Moreover, the “black box” nature of the AI algorithm, especially the deep learning model, may require considerable effort to convince the regulatory authority to approve for its routine use. The liability and indemnity issues related to the manufacturers of the AI system also need to be resolved. Hence, there are still considerable obstacles to overcome before the application of AI-assisted colonoscopy becomes widespread in daily practice.
An externally validated AI system could be one of the promising solutions to increase adenoma detection and to minimize missed lesions during real-time colonoscopy. As of yet, means to ensure adequate mucosal exposure, such as add-on devices and optimal bowel preparation, are also critical in reducing the polyp miss rate in daily colonoscopy practice. Long-term data are also needed to determine the actual clinical benefits of this emerging technology in the reduction of PCCRC.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and Hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Coghlan E S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T, Forouzanfar MH, Hancock J, Hay RJ, Hunter-Merrill R, Huynh C, Hosgood HD, Johnson CO, Jonas JB, Khubchandani J, Kumar GA, Kutz M, Lan Q, Larson HJ, Liang X, Lim SS, Lopez AD, MacIntyre MF, Marczak L, Marquez N, Mokdad AH, Pinho C, Pourmalek F, Salomon JA, Sanabria JR, Sandar L, Sartorius B, Schwartz SM, Shackelford KA, Shibuya K, Stanaway J, Steiner C, Sun J, Takahashi K, Vollset SE, Vos T, Wagner JA, Wang H, Westerman R, Zeeb H, Zoeckler L, Abd-Allah F, Ahmed MB, Alabed S, Alam NK, Aldhahri SF, Alem G, Alemayohu MA, Ali R, Al-Raddadi R, Amare A, Amoako Y, Artaman A, Asayesh H, Atnafu N, Awasthi A, Saleem HB, Barac A, Bedi N, Bensenor I, Berhane A, Bernabé E, Betsu B, Binagwaho A, Boneya D, Campos-Nonato I, Castañeda-Orjuela C, Catalá-López F, Chiang P, Chibueze C, Chitheer A, Choi JY, Cowie B, Damtew S, das Neves J, Dey S, Dharmaratne S, Dhillon P, Ding E, Driscoll T, Ekwueme D, Endries AY, Farvid M, Farzadfar F, Fernandes J, Fischer F, G/Hiwot TT, Gebru A, Gopalani S, Hailu A, Horino M, Horita N, Husseini A, Huybrechts I, Inoue M, Islami F, Jakovljevic M, James S, Javanbakht M, Jee SH, Kasaeian A, Kedir MS, Khader YS, Khang YH, Kim D, Leigh J, Linn S, Lunevicius R, El Razek HMA, Malekzadeh R, Malta DC, Marcenes W, Markos D, Melaku YA, Meles KG, Mendoza W, Mengiste DT, Meretoja TJ, Miller TR, Mohammad KA, Mohammadi A, Mohammed S, Moradi-Lakeh M, Nagel G, Nand D, Le Nguyen Q, Nolte S, Ogbo FA, Oladimeji KE, Oren E, Pa M, Park EK, Pereira DM, Plass D, Qorbani M, Radfar A, Rafay A, Rahman M, Rana SM, Søreide K, Satpathy M, Sawhney M, Sepanlou SG, Shaikh MA, She J, Shiue I, Shore HR, Shrime MG, So S, Soneji S, Stathopoulou V, Stroumpoulis K, Sufiyan MB, Sykes BL, Tabarés-Seisdedos R, Tadese F, Tedla BA, Tessema GA, Thakur JS, Tran BX, Ukwaja KN, Uzochukwu BSC, Vlassov VV, Weiderpass E, Wubshet Terefe M, Yebyo HG, Yimam HH, Yonemoto N, Younis MZ, Yu C, Zaidi Z, Zaki MES, Zenebe ZM, Murray CJL, Naghavi M. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2838] [Cited by in RCA: 2939] [Article Influence: 367.4] [Reference Citation Analysis (0)] |
| 2. | Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2271] [Article Influence: 174.7] [Reference Citation Analysis (1)] |
| 3. | Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 968] [Cited by in RCA: 1147] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 4. | Cheung KS, Chen L, Seto WK, Leung WK. Epidemiology, characteristics, and survival of post-colonoscopy colorectal cancer in Asia: A population-based study. J Gastroenterol Hepatol. 2019;34:1545-1553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 5. | Robertson DJ, Lieberman DA, Winawer SJ, Ahnen DJ, Baron JA, Schatzkin A, Cross AJ, Zauber AG, Church TR, Lance P, Greenberg ER, Martínez ME. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut. 2014;63:949-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 332] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 6. | le Clercq CM, Bouwens MW, Rondagh EJ, Bakker CM, Keulen ET, de Ridder RJ, Winkens B, Masclee AA, Sanduleanu S. Postcolonoscopy colorectal cancers are preventable: a population-based study. Gut. 2014;63:957-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 7. | Sanduleanu S, le Clercq CM, Dekker E, Meijer GA, Rabeneck L, Rutter MD, Valori R, Young GP, Schoen RE; Expert Working Group on ‘Right-sided lesions and interval cancers’, Colorectal Cancer Screening Committee, World Endoscopy Organization. Definition and taxonomy of interval colorectal cancers: a proposal for standardising nomenclature. Gut. 2015;64:1257-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 147] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 8. | Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology. 2019;156:1661-1674.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 360] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 9. | Gupta N. How to Improve Your Adenoma Detection Rate During Colonoscopy. Gastroenterology. 2016;151:1054-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Mahmud N, Cohen J, Tsourides K, Berzin TM. Computer vision and augmented reality in gastrointestinal endoscopy. Gastroenterol Rep (Oxf). 2015;3:179-184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 11. | Wang W, Xu L, Bao Z, Sun L, Hu C, Zhou F, Xu L, Shi D. Differences with experienced nurse assistance during colonoscopy in detecting polyp and adenoma: a randomized clinical trial. Int J Colorectal Dis. 2018;33:561-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Buchner AM, Shahid MW, Heckman MG, Diehl NN, McNeil RB, Cleveland P, Gill KR, Schore A, Ghabril M, Raimondo M, Gross SA, Wallace MB. Trainee participation is associated with increased small adenoma detection. Gastrointest Endosc. 2011;73:1223-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Aslanian HR, Shieh FK, Chan FW, Ciarleglio MM, Deng Y, Rogart JN, Jamidar PA, Siddiqui UD. Nurse observation during colonoscopy increases polyp detection: a randomized prospective study. Am J Gastroenterol. 2013;108:166-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 14. | Bisschops R, East JE, Hassan C, Hazewinkel Y, Kamiński MF, Neumann H, Pellisé M, Antonelli G, Bustamante Balen M, Coron E, Cortas G, Iacucci M, Yuichi M, Longcroft-Wheaton G, Mouzyka S, Pilonis N, Puig I, van Hooft JE, Dekker E. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy (ESGE) Guideline - Update 2019. Endoscopy. 2019;51:1155-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 223] [Article Influence: 37.2] [Reference Citation Analysis (1)] |
| 15. | Clark BT, Rustagi T, Laine L. What level of bowel prep quality requires early repeat colonoscopy: systematic review and meta-analysis of the impact of preparation quality on adenoma detection rate. Am J Gastroenterol. 2014;109:1714-23; quiz 1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 194] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 16. | Barclay RL, Vicari JJ, Doughty AS, Johanson JF, Greenlaw RL. Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. N Engl J Med. 2006;355:2533-2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 943] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 17. | Barclay RI, Vicari JJ, Johanson JF, Greenlaw RI. Variation in adenoma detection rates and colonoscopic withdrawal times during screening colonoscopy. Gastrointest Endosc. 2005;61:Ab107. [DOI] [Full Text] |
| 18. | Barclay RL, Vicari JJ, Greenlaw RL. Effect of a time-dependent colonoscopic withdrawal protocol on adenoma detection during screening colonoscopy. Clin Gastroenterol Hepatol. 2008;6:1091-1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 198] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 19. | Lee HS, Jeon SW, Park HY, Yeo SJ. Improved detection of right colon adenomas with additional retroflexion following two forward-view examinations: a prospective study. Endoscopy. 2017;49:334-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Cohen J, Grunwald D, Grossberg LB, Sawhney MS. The Effect of Right Colon Retroflexion on Adenoma Detection: A Systematic Review and Meta-analysis. J Clin Gastroenterol. 2017;51:818-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. | Cadoni S, Falt P, Rondonotti E, Radaelli F, Fojtik P, Gallittu P, Liggi M, Amato A, Paggi S, Smajstrla V, Urban O, Erriu M, Koo M, Leung FW. Water exchange for screening colonoscopy increases adenoma detection rate: a multicenter, double-blinded, randomized controlled trial. Endoscopy. 2017;49:456-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 22. | Lee CK, Park DI, Lee SH, Hwangbo Y, Eun CS, Han DS, Cha JM, Lee BI, Shin JE. Participation by experienced endoscopy nurses increases the detection rate of colon polyps during a screening colonoscopy: a multicenter, prospective, randomized study. Gastrointest Endosc. 2011;74:1094-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Kudo T, Saito Y, Ikematsu H, Hotta K, Takeuchi Y, Shimatani M, Kawakami K, Tamai N, Mori Y, Maeda Y, Yamada M, Sakamoto T, Matsuda T, Imai K, Ito S, Hamada K, Fukata N, Inoue T, Tajiri H, Yoshimura K, Ishikawa H, Kudo SE. New-generation full-spectrum endoscopy versus standard forward-viewing colonoscopy: a multicenter, randomized, tandem colonoscopy trial (J-FUSE Study). Gastrointest Endosc. 2018;88:854-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Atkinson NSS, Ket S, Bassett P, Aponte D, De Aguiar S, Gupta N, Horimatsu T, Ikematsu H, Inoue T, Kaltenbach T, Leung WK, Matsuda T, Paggi S, Radaelli F, Rastogi A, Rex DK, Sabbagh LC, Saito Y, Sano Y, Saracco GM, Saunders BP, Senore C, Soetikno R, Vemulapalli KC, Jairath V, East JE. Narrow-Band Imaging for Detection of Neoplasia at Colonoscopy: A Meta-analysis of Data From Individual Patients in Randomized Controlled Trials. Gastroenterology. 2019;157:462-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 25. | Leung WK, Lo OS, Liu KS, Tong T, But DY, Lam FY, Hsu AS, Wong SY, Seto WK, Hung IF, Law WL. Detection of colorectal adenoma by narrow band imaging (HQ190) vs. high-definition white light colonoscopy: a randomized controlled trial. Am J Gastroenterol. 2014;109:855-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 26. | Leung WK, Guo CG, Ko MKL, To EWP, Mak LY, Tong TSM, Chen LJ, But DYK, Wong SY, Liu KSH, Tsui V, Lam FYF, Lui TKL, Cheung KS, Lo SH, Hung IFN. Linked color imaging versus narrow-band imaging for colorectal polyp detection: a prospective randomized tandem colonoscopy study. Gastrointest Endosc. 2020;91:104-112.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Rameshshanker R, Tsiamoulos Z, Wilson A, Rajendran A, Bassett P, Tekkis P, Saunders BP. Endoscopic cuff-assisted colonoscopy versus cap-assisted colonoscopy in adenoma detection: randomized tandem study-DEtection in Tandem Endocuff Cap Trial (DETECT). Gastrointest Endosc. 2020;91:894-904.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Russell SJ, Norvig P, Davis E. Artificial intelligence: a modern approach. 3rd ed. Upper Saddle River: Prentice Hall; 2010. |
| 29. | Murphy KP. Machine learning: a probabilistic perspective. Cambridge, MA: MIT Press; 2012. |
| 30. | Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge, Massachusetts: The MIT Press; 2016; . |
| 31. | Takiyama H, Ozawa T, Ishihara S, Fujishiro M, Shichijo S, Nomura S, Miura M, Tada T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci Rep. 2018;8:7497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 32. | Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Commun Acm. 2017;60:84-90. [DOI] [Full Text] |
| 33. | Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging. 2016;35:1285-1298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3594] [Cited by in RCA: 1905] [Article Influence: 211.7] [Reference Citation Analysis (0)] |
| 34. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 428] [Article Influence: 61.1] [Reference Citation Analysis (1)] |
| 35. | Figueiredo PN, Figueiredo IN, Pinto L, Kumar S, Tsai YR, Mamonov AV. Polyp detection with computer-aided diagnosis in white light colonoscopy: comparison of three different methods. Endosc Int Open. 2019;7:E209-E215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta-analysis. Gastrointest Endosc. 2020;92:11-22.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 37. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 542] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 38. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 291] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 39. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 385] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 40. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 256] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 41. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 203] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 42. | Lui TK, Hui CK, Tsui VW, Cheung KS, Ko MK, aCC Foo D, Mak LY, Yeung CK, Lui TH, Wong SY, Leung WK. New insights on missed colonic lesions during colonoscopy through artificial intelligence-assisted real-time detection (with video). Gastrointest Endosc. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 43. | Wang P, Liu P, Glissen Brown JR, Berzin TM, Zhou G, Lei S, Liu X, Li L, Xiao X. Lower Adenoma Miss Rate of Computer-aided Detection-Assisted Colonoscopy vs Routine White-Light Colonoscopy in a Prospective Tandem Study. Gastroenterology. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 44. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 408] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 45. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 273] [Article Influence: 39.0] [Reference Citation Analysis (0)] |