Published online Aug 21, 2018. doi: 10.3748/wjg.v24.i31.3488
Peer-review started: May 4, 2018
First decision: May 23, 2018
Revised: June 1, 2018
Accepted: June 25, 2018
Article in press: June 25, 2018
Published online: August 21, 2018
Hepatitis B virus (HBV) infection is a global public health concern. HBV causes chronic infection in patients and can lead to liver cirrhosis, hepatocellular carcinoma, and other severe liver diseases. Thus, understanding HBV-related pathogenesis is of particular importance for prevention and clinical intervention. HBV surface antigens are indispensable for HBV virion formation and are useful viral markers for diagnosis and clinical assessment. During chronic HBV infection, HBV genomes may acquire and accumulate mutations and deletions, leading to the expression of defective HBV surface antigens. These defective HBV surface antigens have been found to play important roles in the progression of HBV-associated liver diseases. In this review, we focus our discussion on the nature of defective HBV surface antigen mutations and their contribution to the pathogenesis of fulminant hepatitis B. The relationship between defective surface antigens and occult HBV infection are also discussed.
Core tip: Defective surface antigen mutation is a type of mutation with great clinical relevance. Many previous publications have explored the association of defective surface antigen mutation with the development of hepatitis B virus (HBV)-associated hepatocellular carcinoma. However, there are no reviews available that elaborate on the relationship between defective surface antigen mutation and HBV-associated fulminant hepatitis (FH), as well as occult hepatitis B virus infection (OBI). This review will focus on these two aspects to discuss the nature of defective HBV surface antigen mutations and their contribution to the pathogenesis of FH. The relationship between defective surface antigens and OBI are also discussed.
- Citation: Wu CC, Chen YS, Cao L, Chen XW, Lu MJ. Hepatitis B virus infection: Defective surface antigen expression and pathogenesis. World J Gastroenterol 2018; 24(31): 3488-3499
- URL: https://www.wjgnet.com/1007-9327/full/v24/i31/3488.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i31.3488
Hepatitis B virus (HBV) is an important human pathogen that has caused chronic infections worldwide[1]. Recent data obtained from a modeling study has shown that the global prevalence of hepatitis B surface antigen (HBsAg) was 3.9% in 2016, corresponding to an estimated 290 million infections worldwide[2]. HBV mainly infects hepatocytes and causes a wide spectrum of clinical manifestations, ranging from an asymptomatic carrier state to acute or chronic hepatitis, with progression to liver cirrhosis, hepatocellular carcinoma (HCC), and other severe liver diseases[3,4]. Currently, interferon-α and nucleotide analogs are used to treat chronic HBV (CHB) infections; however, the outcome is far from satisfactory[5,6]. Prophylaxis using the current HBV vaccines has no impact on existing infections. Therapeutic vaccines of chronic HBV infection are under investigation, but further development is still required[7]. Therefore, understanding the molecular pathogenesis of HBV infection will provide opportunities for the development of better therapies and vaccines.
HBV belongs to the family Hepadnaviridae and is a small, enveloped virus with a partially double-stranded DNA genome approximately 3.2 kb in size[8]. During the life cycle of HBV, pre-genomic RNA (pgRNA) is transcribed from covalently closed circular DNA (cccDNA) and serves as the template for HBV DNA replication through a viral polymerase-mediated reverse transcription[9,10]. Because viral polymerase lacks a proof-reading function, the HBV genome evolves with an estimated rate of nucleotide substitutions of 1 × 10-3 to 1 × 10-6 per replication cycle, according to various investigators[11]. Although HBV genome replication involves a step of reverse transcription, which is similar to retroviral replication, the complex HBV genome structure with overlapping open reading frames and regulatory sequences apparently limits the spectrum and rate of mutations[3,12]. Nevertheless, this unique replication strategy leads to the great diversity of HBV genomes, thus resulting in the occurrence of various genotypes, subtypes, mutants, recombinants, and even viral quasi-species in the context of long-term HBV evolution[13,14]. Several reports have suggested that the emergence of HBV variants plays important roles in the progression of HBV-associated liver diseases[11,15-18]. Defective surface antigen mutation is a type of mutation with great clinical relevance[11,15,19]. In this review, we report the current information on HBV surface antigen mutations. Further, we focus our discussion on the contribution of defective surface antigen mutations on the pathogenesis of HBV-associated liver diseases.
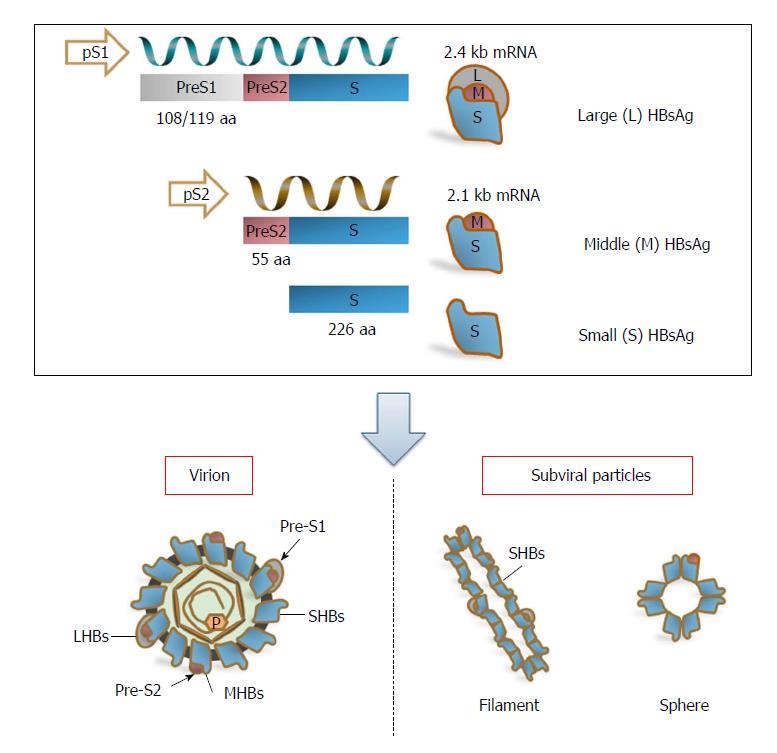
Three viral envelope/surface proteins - large surface antigens (LHBs), middle surface antigens (MHBs), and small surface antigens (SHBs) - are expressed from a single open reading frame (S-ORF)[20,21], but they are translated from two different mRNAs. LHBs are encoded by the 2.4 kb subgenomic RNA, and MHBs and SHBs are encoded by the 2.1 kb subgenomic RNA[3]. Subgenomic RNAs of 2.4 kb and 2.1 kb are driven by preS1 and preS2/S promoters, respectively, allowing variable regulation of protein expression[3]. The preS1 promoter is situated within the upstream region of the S-ORF, whereas the preS2 promoter corresponds to the preS1 domain[21]. Therefore, the transcription of the 2.1 kb subgenomic RNA is also regulated by the preS1 domain[11] (Figure 1).
The three surface proteins share the same carboxy-terminal region and only differ in length due to their amino-terminal regions. As a result, the LHBs contain the preS1 + preS2 + S [389 or 400 amino acid (aa) residues], MHBs contain the preS2 + S (281 aa residues), and SHBs contain the S domain (226 aa residues) alone[3,20,22] (Figure 1). Additionally, a truncated and mutated preS2/S (the LHBs and truncated MHBs) can be produced by integrated viral sequences that are defective for replication[23,24]. LHBs, MHBs, and SHBs are important for HBV structure and life cycle. Besides mediating HBV entry through binding to HBV receptors, the sodium taurocholate co-transporting polypeptide (NTCP) on hepatocytes, via the preS1 2-48 aa domain (numbering for HBV-genotype D) and subsequent infection, LHBs are indispensable for the formation and budding of virions[3,25-29]. It has been proposed that LHBs rearrange their structure during the maturation of HBV virions and thereby regulate the release and infectivity of virions[30-32]. The exact role of MHBs in the HBV life cycle remains an enigma. Early reports indicated that MHBs might be dispensable for HBV replication and virion formation; however, our data and those of other groups have shown that MHBs play a role in virion secretion[33-36]. Recently, MHBs were found to interact with ceruloplasmin and influence the production of extracellular virions[34]. As the predominant component of viral particles, including infectious virions and noninfectious subviral particles, SHBs are necessary for the production of virions and subviral particles[35]. For mature/infectious virions, LHBs, MHBs, and SHBs are present in the envelopes at a ratio of approximately 1:1:4[20]. Disturbance of this proportion impairs the production and release of virions[33]. For subviral particles, their amount outnumbers virions by 10000- to 1000000-fold, and the particles are detected serologically as HBsAg[11,37]. The secretion of subviral particles can also be suppressed by LHBs in a dose-dependent manner[38-41], thus promoting the S protein toward virion formation.
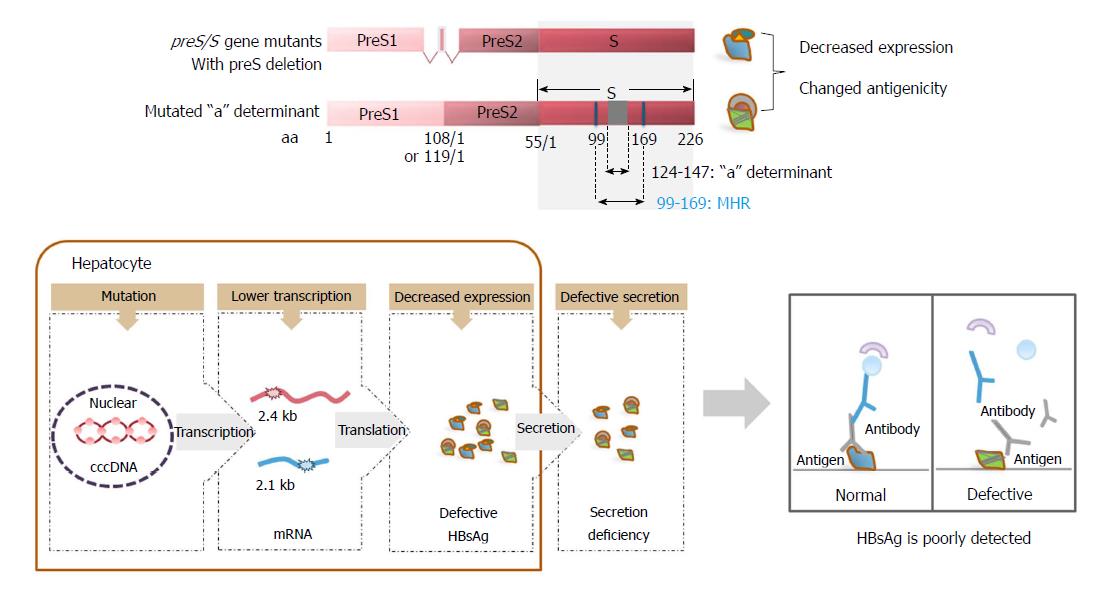
In addition, preS1, preS2, and S domains contain various B- and T-cell epitopes, which play an important role in inducing the host immune response[42,43]. The major hydrophilic region (MHR) between aa 100-169 of SHBs, especially the a-determinant located at aa 124-147, serves as the most important antigenic determinant in HBV surface proteins and is essential for HBsAg detection and HBV vaccine development[44,45]. Plasma-derived and recombinant HBsAg have been used for vaccine preparations and have induced strong specific and protective antibody responses in vaccines[46-48]. The presence of anti-HBs antibodies is considered to confer immunity against HBV infection. In contrast, a high quantity of circulating HBsAg in chronically HBV-infected patients is proposed as a factor leading to immune disturbance. Defective peripheral HBsAg-specific T cell responses in chronically infected patients were found to be correlated with serum HBsAg titers[49,50], suggesting that HBsAg overproduction influences the host’s immune system in a way that is advantageous for the virus. In vitro, HBsAg can interfere with Toll-like receptor functions and trigger interleukin (IL)-10 production in Kuppfer cells[51-54]. Recently, published data has suggested that HBsAg may facilitate the induction of myeloid-derived suppressor cells in chronically HBV-infected patients[55]. HBsAg is also associated with the induction of regulatory T cells, as shown in HBV mouse models[56,57]. Thus, HBsAg is not only a structural component of virions and subviral particles, but it also serves as an important immune modulator.
HBsAg mutants were first identified in individuals vaccinated against HBV but who were infected despite the presence of protective anti-HBs antibodies[58]. Those “immune escape” mutants with aa substitutions within a-determinants were found in different clinical settings, including vaccines, transplant patients receiving hyperimmunoglobulins, and immunocompromised patients with HBV reactivation[59-61]. Such mutant HBsAg commonly showed reduced binding to anti-HBs antibodies and decreased reactivity in established HBsAg detection assays[59-65]. The most widely known mutation is the sG145R mutation, which has been shown to be replication competent, may persist stably over time, and may be transmitted vertically or horizontally[66-69]. The sG145R mutation induces a strongly impaired anti-HBs antibody response, which could not efficiently clear HBsAg in an HBV hydrodynamic injection mouse model[70]. A similar result was also observed for another immune escape mutation, sK122I, indicating that such a defective surface antigen mutation may impair HBsAg neutralization and clearance during HBV infection. In addition, sG145R, sK122I, and other immune escape mutants occurring in the a-determinant of SHBs, such as the sT123N mutation, could affect HBsAg secretion[70-73].
Recently, chronically HBV-infected patients routinely received antiviral therapy based on nucleotide analogs[74]. Treatment with first-generation drugs, such as famciclovir and adeforvir, resulted in the emergence of drug-resistant HBV mutants, with aa substitutions within the HBV polymerase domain[75]. Some drug-resistant mutations occurring in the viral polymerase may lead to a stop codon mutation in the overlapping surface gene, cause intracellular retention of surface proteins, and result in secretion defects of viral particles, such as rtA181T/sW172*, rtM204I/sW196*, and rtV191I/sW182*, as shown in previous reports[76-78] and in our unpublished data. The primary sW182* mutation has also been identified in CHB patients. It was found to induce retention of the truncated S protein in the perinuclear endoplasmic reticulum (ER) and was associated with lower HBV transcript levels owing to decreased stability, but without impact on HBV replication[79].
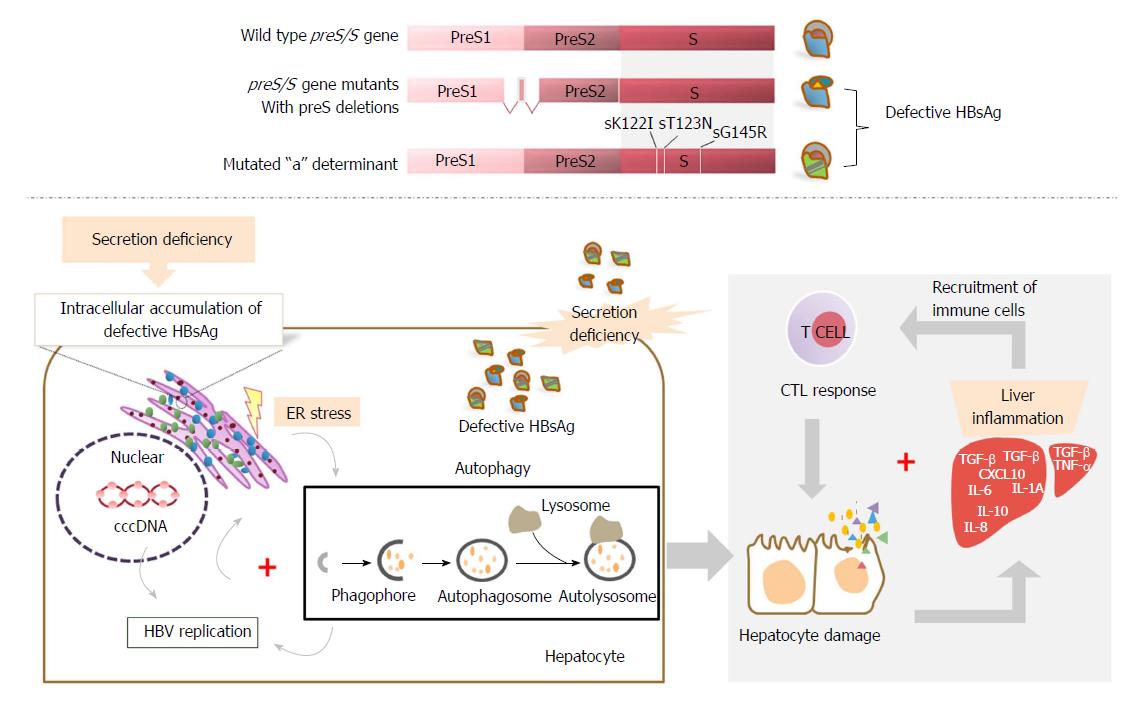
Defective surface antigen mutations have been frequently detected in chronic HBV infection[16,71-73,80,81], in which deletions in the preS domains are the most common mutations[80,81]. Deletions in the preS domains are often clustered at the 3’ end of preS1 and the 5’ end of preS2[11,19,81-83]. Given that the preS2/S promoter is situated within the preS1 domain[11], deletions at the 3’ end of the preS1 may reduce MHBs and SHBs expression at the transcriptional level. Deletions at the 5’ end of the preS2 may remove the N-terminal preS2 domain, including the start codon of preS2 in the MHBs protein, leading to an impaired or complete loss of MHBs expression[84]. These changes may disrupt the proper LHBs, MHBs, and SHBs ratio in the envelopes of virions. In addition, the junction between the preS1 and preS2 domain is required for virion formation[32]. For these reasons, preS deletions may potentially affect virion assembly, stability, or infectivity.
A large amount of evidence has demonstrated that DHBV envelope proteins can regulate cccDNA formation and amplification[85,86]. Infection of envelope protein-deficient recombinant DHBV results in more cccDNA accumulation[85,87,88]. Similarly, deficiencies in HBV envelope proteins can modestly increase the cccDNA level and result in a dramatic accumulation of deproteinized rcDNA[89-91]. It has been demonstrated that preS/S mutants with surface antigen secretion deficiency isolated from patients can lead to an increased accumulation of cccDNA molecules in the nuclei[79]. Therefore, defective surface antigen mutation may affect cccDNA synthesis and amplification.
Defective surface antigen mutations have been found in acute hepatitis B infection, chronic hepatitis B infection, and occult HBV infection and are associated with advanced liver disease, including liver cirrhosis, fulminant hepatitis B, and HCC[15,82,92-105]. It has been questioned whether HBV mutants arise due to viral adaptation to inflammation and decreased liver function or, alternatively, causally contribute to liver pathogenesis. The mechanism of defective surface antigen mutations to HCC development has been widely elucidated[11,23,41,106-111]. Here, we will emphasize in our discussion the relationship between defective surface antigen mutations and fulminant hepatitis B, as well as occult HBV infection.
There is increasing evidence that defective surface antigen expression may play a role in the pathogenesis of fulminant hepatitis (FH). preS deletions, particularly those unable to synthesize the MHBs protein, have been associated with cases of FH[16,71,112,113], suggesting the potential pathogenic role of preS deletions. A mutation in the CAAT element of the S promoter has been found in the HBV genome isolated from an FH patient. This mutation led to excessive LHBs expression over MHBs and SHBs proteins and resulted in virus retention and misassembly[114-116]. Obviously, the accumulation of LHBs may be due to hepatocyte injury, as shown in transgenic mice with LHBs expression[41]. One of our previous studies also identified deletions within the preS regions from HBV strains isolated from a patient with HBV-associated FH[84]. In addition, a hepatitis B immune globulin (HBIG)-escape mutant sG145R on the HBsAg, causing 30% inhibition of virion secretion, has been identified from a study on FH strains, suggesting the potential role of defective surface antigen expression in the fulminant clinical course of HBV infection[71].
Mechanistically, defective surface antigen expression, such as specific mutations in the preS/S gene, may lead to secretion defects of viral proteins and particles, resulting in an accumulation of viral products in the ER of hepatocytes and causing ER stress and hepatocyte injury[16]. Subsequently, autophagy may be triggered[117-125] and thus enhance HBV replication[126,127]. Consistent with this speculation, it has been demonstrated that defective surface antigen expression may increase the replication capability of HBV, albeit the mechanism is still undefined[71,84]. In addition, the deficiency of hepadnavirus envelope proteins can result in accumulation of cccDNA[85,87,88] or deproteinized rcDNA[89-91] and may ultimately cause death of the infected hepatocytes by a direct cytopathic effect[85,87,88]. Meanwhile, the increase of the cccDNA level may facilitate HBV replication. Both the defect in viral particle secretion and enhanced replication competence may contribute to the severity of fulminant hepatitis[128].
The adaptive immune response, particularly the cytotoxic T lymphocyte (CTL) response, plays a crucial role in viral clearance and disease pathogenesis of HBV infection[129-131]. Intracellular retention of HBV surface proteins was found to be associated with FH in a transgenic mouse model showing panlobular necrosis and hepatic failure by inducing the indirect cytotoxic activity of CTLs[132]. In this setting, intracellular accumulation of viral products due to defective surface antigen expression mutations may cause liver damage through abnormal activation of the CTL response. Consistently, we also observed significantly stronger intrahepatic CTL responses and antibody responses specific to secretion-deficient HBsAg due to preS deletions[84]. A preS deletion mutant was found in a patient with acute exacerbation of liver diseases, along with wild-type HBV genomes. The co-existence of deletion mutants and wild-type HBV apparently allows the complementation and enhancement of HBV genome replication in hepatocytes. In an HBV mouse model, co-replication of a deletion mutant and wild-type HBV induced higher cellular and humoral immunity. Our findings further suggested the proposed role of HBV variants in the immunopathogenesis of HBV infection. Moreover, the mutations associated with defective surface antigen expression, such as deletion or missense mutation of the preS2 ATG codon, can cause deletions or alterations of B- and T-cell epitopes located in preS1 and preS2 proteins. Considering that M protein-specific T- and B-cell immunities are important early events in the host immune response to HBV infection[43], these mutations may lead to an immune evasion and thus likely favor a more severe clinical course of infection[14,133]. In chronic HBV infection, high HBV replication levels were found to be associated with lower cellular immune responses to HBV; however, massive infiltration of unspecific immune cells occurred within the liver, accompanied by severe liver damage[134-136]. Thus, the presence of these mutations, including aa substitutions at the immunodominant epitopes for B or T cell recognition, may contribute to the spread of highly replicative escape mutants. It may also facilitate the development of fulminant hepatitis in chronically HBV-infected patients and heavily immunocompromised patients, like those with human immunodeficiency virus (HIV) co-infection[137] (Figure 2).
Occult HBV infection (OBI) is characterized by the presence of very low levels of HBV DNA in the plasma and/or liver of individuals negative for HBV surface antigen (HBsAg) and positive/negative for antibodies to the hepatitis core antigen (anti-HBc)[45,138,139]. OBI harbors the potential risk of HBV transmission through blood transfusion, organ transplantation, and hemodialysis as well as from occult infection or HBsAg-positive mothers to newborns[45]. The persistence of OBI may lead to the development of cirrhosis and HCC[45,140-145]. The reactivation of OBI can occur in patients following chemotherapy, immunosuppressive therapy, and after transplantation as well as in patients co-infected with HIV or hepatitis C virus (HCV)[45,146,147], which can result in the development of fulminant hepatitis and death[139,148-153].
Defective surface antigen expression mutations may be associated with OBI. Point mutations and deletions as well as insertion mutations are commonly encountered in OBI, in which mutations in the preS/S gene are the most extensively studied[45]. High frequencies of MHR mutations, including those mutations within and outside of the a-determinant, have been observed in OBI strains of individuals[154-158]. In vitro and in vivo experiments have demonstrated that these MHR mutations can significantly decrease the detection sensitivity of commercial HBsAg immunoassays and impair virion and/or S protein secretion[156]. preS/S mutations with deletions covering the preS1 and preS2/S promoters, preS1 region, and preS2 region have been frequently reported in OBI. This can alter the transcription of 2.4 kb and 2.1 kb HBV RNAs, expression of three envelope proteins, and the ratio of LHBs/MHBs/SHBs proteins[45]. preS/S insertions, such as 2-8 aa insertions between codons 121 and 124 located upstream of the a-determinant, have also been observed in OBI patients[159].
On one hand, these mutations associated with defective surface antigen expression can directly decrease the levels of surface antigens. On the other hand, these mutations can cause the retention and accumulation of HBsAg within cells and impair the secretion of HBsAg by altering the ratio of LHBs/MHBs/SHBs proteins[72,73,160,161]. Therefore, circulating HBsAg levels are low in the peripheral blood. Moreover, it is well documented that neutralizing antibodies produced during natural infection, or following active or passive immunization against HBV, are targeted to the conformational epitopes of the a-determinant[162]. Hence, single or multiple mutations occurring within this region can lead to conformational changes with impaired antigenicity[72,160]. A recent report has identified novel SHBs mutations outside the MHR from untreated CHB patients. These mutations impaired virion secretion and caused lower binding affinity to antibodies used for HBsAg immunoassays[163]. For these reasons, the mutations can render HBsAg undetectable or poorly detected by immunoassays based on monoclonal antibodies against wild-type virus[60,62,65,164], contributing to some cases of OBI[165-170] (Figure 3).
Defective surface antigen expression has been well documented to be relevant for the progression of HBV-associated liver diseases, such as HCC. However, the role of defective surface antigen expression in FH still needs to be clarified in future research, particularly, using in vivo models and in patients. The exact molecular mechanisms of how defective HBV surface antigens cause damage to hepatocytes and induce liver injury and subsequent pathogenic processes should be investigated. A deep understanding of the molecular mechanisms of HBV pathogenesis related to defective surface antigens is crucial to designing future therapeutic approaches. A critical question would be whether currently used nucleotide analogues (NAs) and interferon-based therapies can prevent such pathogenic processes. NAs are able to efficiently inhibit HBV DNA synthesis but not gene expression. Thus, HBV proteins, including surface antigens, are continuously produced under NA therapies. Another problem is the production of mutated HBV proteins from integrated HBV DNA, which are not controlled by NA therapies at all. Thus, specific interventions may be required to block the pathogenic potential of HBV proteins, besides efficient inhibition of HBV DNA synthesis. RNA silencing may be a suitable choice to achieve this goal[5,6,171,172].
An additional issue to be addressed is whether the defective surface antigen-related mutations may represent novel biomarkers of OBI. With improvement of HBV antigen and DNA detection assays, OBI will likely be easier to diagnose in the future. However, the question remains whether OBI may be related to significant HBV pathogenesis and require therapeutic interventions, such as prophylaxis and antiviral therapy, to prevent HBV reactivation[173].
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Farshadpour F, Zhao HT S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Huang Y
| 1. | Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212-2219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1217] [Cited by in F6Publishing: 1250] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 2. | Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 961] [Cited by in F6Publishing: 1060] [Article Influence: 176.7] [Reference Citation Analysis (1)] |
| 3. | Glebe D, Bremer CM. The molecular virology of hepatitis B virus. Semin Liver Dis. 2013;33:103-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 4. | Bernal W, Auzinger G, Dhawan A, Wendon J. Acute liver failure. Lancet. 2010;376:190-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 720] [Cited by in F6Publishing: 677] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 5. | Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: From discovery to regulatory approval. Hepatology. 2017;66:1296-1313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 203] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 6. | Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: From discovery to regulatory approval. J Hepatol. 2017;67:847-861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 155] [Article Influence: 22.1] [Reference Citation Analysis (1)] |
| 7. | Kosinska AD, Liu J, Lu M, Roggendorf M. Therapeutic vaccination and immunomodulation in the treatment of chronic hepatitis B: preclinical studies in the woodchuck. Med Microbiol Immunol. 2015;204:103-114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 569] [Cited by in F6Publishing: 556] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 9. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 548] [Cited by in F6Publishing: 589] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 10. | Seeger C. Control of viral transcripts as a concept for future HBV therapies. Curr Opin Virol. 2018;30:18-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4-S16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 270] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 12. | Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J. 2013;10:239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 13. | Lin CL, Kao JH. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5:a021436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 150] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 14. | Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 201] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 15. | Kao JH, Chen PJ, Chen DS. Recent advances in the research of hepatitis B virus-related hepatocellular carcinoma: epidemiologic and molecular biological aspects. Adv Cancer Res. 2010;108:21-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61:408-417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 186] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 17. | Shen T, Yan XM. Hepatitis B virus genetic mutations and evolution in liver diseases. World J Gastroenterol. 2014;20:5435-5441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Zhang ZH, Wu CC, Chen XW, Li X, Li J, Lu MJ. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J Gastroenterol. 2016;22:126-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 68] [Cited by in F6Publishing: 65] [Article Influence: 8.1] [Reference Citation Analysis (1)] |
| 19. | Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153-1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 231] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 20. | Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol. 1984;52:396-402. [PubMed] [Cited in This Article: ] |
| 21. | Cattaneo R, Will H, Hernandez N, Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983;305:336-338. [PubMed] [Cited in This Article: ] |
| 22. | Stibbe W, Gerlich WH. Structural relationships between minor and major proteins of hepatitis B surface antigen. J Virol. 1983;46:626-628. [PubMed] [Cited in This Article: ] |
| 23. | Schlüter V, Meyer M, Hofschneider PH, Koshy R, Caselmann WH. Integrated hepatitis B virus X and 3’ truncated preS/S sequences derived from human hepatomas encode functionally active transactivators. Oncogene. 1994;9:3335-3344. [PubMed] [Cited in This Article: ] |
| 24. | Li YW, Yang FC, Lu HQ, Zhang JS. Hepatocellular carcinoma and hepatitis B surface protein. World J Gastroenterol. 2016;22:1943-1952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 40] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 25. | Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol. 1999;73:2052-2057. [PubMed] [Cited in This Article: ] |
| 26. | Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759-1768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 322] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 27. | Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grün S, Bulavaite A, Sasnauskas K, Gerlich WH. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234-245. [PubMed] [Cited in This Article: ] |
| 28. | Glebe D, Aliakbari M, Krass P, Knoop EV, Valerius KP, Gerlich WH. Pre-s1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J Virol. 2003;77:9511-9521. [PubMed] [Cited in This Article: ] |
| 29. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1280] [Cited by in F6Publishing: 1428] [Article Influence: 119.0] [Reference Citation Analysis (0)] |
| 30. | Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048-1057. [PubMed] [Cited in This Article: ] |
| 31. | Bruss V, Lu X, Thomssen R, Gerlich WH. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273-2279. [PubMed] [Cited in This Article: ] |
| 32. | Bruss V. Hepatitis B virus morphogenesis. World J Gastroenterol. 2007;13:65-73. [PubMed] [Cited in This Article: ] |
| 33. | Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, Tong S. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol. 2009;83:11152-11165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 34. | Zhao K, Wu C, Yao Y, Cao L, Zhang Z, Yuan Y, Wang Y, Pei R, Chen J, Hu X. Ceruloplasmin inhibits the production of extracellular hepatitis B virions by targeting its middle surface protein. J Gen Virol. 2017;98:1410-1421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059-1063. [PubMed] [Cited in This Article: ] |
| 36. | Fernholz D, Galle PR, Stemler M, Brunetto M, Bonino F, Will H. Infectious hepatitis B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology. 1993;194:137-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 110] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 37. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1700] [Cited by in F6Publishing: 1651] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 38. | Persing DH, Varmus HE, Ganem D. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science. 1986;234:1388-1391. [PubMed] [Cited in This Article: ] |
| 39. | Ou JH, Rutter WJ. Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J Virol. 1987;61:782-786. [PubMed] [Cited in This Article: ] |
| 40. | Chisari FV, Filippi P, McLachlan A, Milich DR, Riggs M, Lee S, Palmiter RD, Pinkert CA, Brinster RL. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol. 1986;60:880-887. [PubMed] [Cited in This Article: ] |
| 41. | Chisari FV, Filippi P, Buras J, McLachlan A, Popper H, Pinkert CA, Palmiter RD, Brinster RL. Structural and pathological effects of synthesis of hepatitis B virus large envelope polypeptide in transgenic mice. Proc Natl Acad Sci USA. 1987;84:6909-6913. [PubMed] [Cited in This Article: ] |
| 42. | Nayersina R, Fowler P, Guilhot S, Missale G, Cerny A, Schlicht HJ, Vitiello A, Chesnut R, Person JL, Redeker AG. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J Immunol. 1993;150:4659-4671. [PubMed] [Cited in This Article: ] |
| 43. | Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1189] [Cited by in F6Publishing: 1171] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 44. | Coppola N, Onorato L, Minichini C, Di Caprio G, Starace M, Sagnelli C, Sagnelli E. Clinical significance of hepatitis B surface antigen mutants. World J Hepatol. 2015;7:2729-2739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 63] [Article Influence: 7.0] [Reference Citation Analysis (2)] |
| 45. | Zhu HL, Li X, Li J, Zhang ZH. Genetic variation of occult hepatitis B virus infection. World J Gastroenterol. 2016;22:3531-3546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 52] [Cited by in F6Publishing: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 46. | Maupas P, Chiron JP, Barin F, Coursaget P, Goudeau A, Perrin J, Denis F, Mar ID. Efficacy of hepatitis B vaccine in prevention of early HBsAg carrier state in children. Controlled trial in an endemic area (Senegal). Lancet. 1981;1:289-292. [PubMed] [Cited in This Article: ] |
| 47. | Szmuness W, Stevens CE, Harley EJ, Zang EA, Oleszko WR, William DC, Sadovsky R, Morrison JM, Kellner A. Hepatitis B vaccine: demonstration of efficacy in a controlled clinical trial in a high-risk population in the United States. N Engl J Med. 1980;303:833-841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 791] [Cited by in F6Publishing: 654] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 48. | Valenzuela P, Medina A, Rutter WJ, Ammerer G, Hall BD. Synthesis and assembly of hepatitis B virus surface antigen particles in yeast. Nature. 1982;298:347-350. [PubMed] [Cited in This Article: ] |
| 49. | Reignat S, Webster GJ, Brown D, Ogg GS, King A, Seneviratne SL, Dusheiko G, Williams R, Maini MK, Bertoletti A. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. J Exp Med. 2002;195:1089-1101. [PubMed] [Cited in This Article: ] |
| 50. | Guidotti LG, Isogawa M, Chisari FV. Host-virus interactions in hepatitis B virus infection. Curr Opin Immunol. 2015;36:61-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 51. | Wang S, Chen Z, Hu C, Qian F, Cheng Y, Wu M, Shi B, Chen J, Hu Y, Yuan Z. Hepatitis B virus surface antigen selectively inhibits TLR2 ligand-induced IL-12 production in monocytes/macrophages by interfering with JNK activation. J Immunol. 2013;190:5142-5151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 113] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 52. | Liu J, Yu Q, Wu W, Huang X, Broering R, Werner M, Roggendorf M, Yang D, Lu M. TLR2 Stimulation Strengthens Intrahepatic Myeloid-Derived Cell-Mediated T Cell Tolerance through Inducing Kupffer Cell Expansion and IL-10 Production. J Immunol. 2018;200:2341-2351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 53. | Jiang M, Broering R, Trippler M, Poggenpohl L, Fiedler M, Gerken G, Lu M, Schlaak JF. Toll-like receptor-mediated immune responses are attenuated in the presence of high levels of hepatitis B virus surface antigen. J Viral Hepat. 2014;21:860-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (2)] |
| 54. | Wu J, Meng Z, Jiang M, Pei R, Trippler M, Broering R, Bucchi A, Sowa JP, Dittmer U, Yang D. Hepatitis B virus suppresses toll-like receptor-mediated innate immune responses in murine parenchymal and nonparenchymal liver cells. Hepatology. 2009;49:1132-1140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 268] [Article Influence: 17.9] [Reference Citation Analysis (3)] |
| 55. | Fang Z, Li J, Yu X, Zhang D, Ren G, Shi B, Wang C, Kosinska AD, Wang S, Zhou X. Polarization of Monocytic Myeloid-Derived Suppressor Cells by Hepatitis B Surface Antigen Is Mediated via ERK/IL-6/STAT3 Signaling Feedback and Restrains the Activation of T Cells in Chronic Hepatitis B Virus Infection. J Immunol. 2015;195:4873-4883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 56. | Dietze KK, Schimmer S, Kretzmer F, Wang J, Lin Y, Huang X, Wu W, Wang B, Lu M, Dittmer U. Characterization of the Treg Response in the Hepatitis B Virus Hydrodynamic Injection Mouse Model. PLoS One. 2016;11:e0151717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 57. | Kosinska AD, Pishraft-Sabet L, Wu W, Fang Z, Lenart M, Chen J, Dietze KK, Wang C, Kemper T, Lin Y. Low hepatitis B virus-specific T-cell response in males correlates with high regulatory T-cell numbers in murine models. Hepatology. 2017;66:69-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Zanetti AR, Tanzi E, Manzillo G, Maio G, Sbreglia C, Caporaso N, Thomas H, Zuckerman AJ. Hepatitis B variant in Europe. Lancet. 1988;2:1132-1133. [PubMed] [Cited in This Article: ] |
| 59. | Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325-329. [PubMed] [Cited in This Article: ] |
| 60. | Cooreman MP, van Roosmalen MH, te Morsche R, Sünnen CM, de Ven EM, Jansen JB, Tytgat GN, de Wit PL, Paulij WP. Characterization of the reactivity pattern of murine monoclonal antibodies against wild-type hepatitis B surface antigen to G145R and other naturally occurring “a” loop escape mutations. Hepatology. 1999;30:1287-1292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 62. | Chiou HL, Lee TS, Kuo J, Mau YC, Ho MS. Altered antigenicity of ‘a’ determinant variants of hepatitis B virus. J Gen Virol. 1997;78:2639-2645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 63. | Günther S, Fischer L, Pult I, Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res. 1999;52:25-137. [PubMed] [Cited in This Article: ] |
| 64. | Lu M, Lorentz T. De novo infection in a renal transplant recipient caused by novel mutants of hepatitis B virus despite the presence of protective anti-hepatitis B surface antibody. J Infect Dis. 2003;187:1323-1326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 65. | Seddigh-Tonekaboni S, Waters JA, Jeffers S, Gehrke R, Ofenloch B, Horsch A, Hess G, Thomas HC, Karayiannis P. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J Med Virol. 2000;60:113-121. [PubMed] [Cited in This Article: ] |
| 66. | Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, Lok AS. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology. 1998;27:213-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 271] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 67. | Hsu HY, Chang MH, Ni YH, Lin HH, Wang SM, Chen DS. Surface gene mutants of hepatitis B virus in infants who develop acute or chronic infections despite immunoprophylaxis. Hepatology. 1997;26:786-791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 141] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 68. | Okamoto H, Yano K, Nozaki Y, Matsui A, Miyazaki H, Yamamoto K, Tsuda F, Machida A, Mishiro S. Mutations within the S gene of hepatitis B virus transmitted from mothers to babies immunized with hepatitis B immune globulin and vaccine. Pediatr Res. 1992;32:264-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 146] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 69. | Protzer-Knolle U, Naumann U, Bartenschlager R, Berg T, Hopf U, Meyer zum Büschenfelde KH, Neuhaus P, Gerken G. Hepatitis B virus with antigenically altered hepatitis B surface antigen is selected by high-dose hepatitis B immune globulin after liver transplantation. Hepatology. 1998;27:254-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 216] [Cited by in F6Publishing: 220] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 70. | Wu C, Deng W, Deng L, Cao L, Qin B, Li S, Wang Y, Pei R, Yang D, Lu M. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol. 2012;86:4658-4669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 71. | Kalinina T, Riu A, Fischer L, Will H, Sterneck M. A dominant hepatitis B virus population defective in virus secretion because of several S-gene mutations from a patient with fulminant hepatitis. Hepatology. 2001;34:385-394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 72. | Wu C, Zhang X, Tian Y, Song J, Yang D, Roggendorf M, Lu M, Chen X. Biological significance of amino acid substitutions in hepatitis B surface antigen (HBsAg) for glycosylation, secretion, antigenicity and immunogenicity of HBsAg and hepatitis B virus replication. J Gen Virol. 2010;91:483-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 73. | Li S, Zhao K, Liu S, Wu C, Yao Y, Cao L, Hu X, Zhou Y, Wang Y, Pei R. HBsAg sT123N mutation induces stronger antibody responses to HBsAg and HBcAg and accelerates in vivo HBsAg clearance. Virus Res. 2015;210:119-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Zoulim F, Durantel D. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb Perspect Med. 2015;5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 117] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 75. | Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593-1608.e1-2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 517] [Cited by in F6Publishing: 515] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 76. | Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology. 2008;48:88-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 191] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 77. | Ahn SH, Park YK, Park ES, Kim JH, Kim DH, Lim KH, Jang MS, Choe WH, Ko SY, Sung IK. The impact of the hepatitis B virus polymerase rtA181T mutation on replication and drug resistance is potentially affected by overlapping changes in surface gene. J Virol. 2014;88:6805-6818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 78. | Yeh CT. Development of HBV S gene mutants in chronic hepatitis B patients receiving nucleotide/nucleoside analogue therapy. Antivir Ther. 2010;15:471-475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 79. | Pollicino T, Amaddeo G, Restuccia A, Raffa G, Alibrandi A, Cutroneo G, Favaloro A, Maimone S, Squadrito G, Raimondo G. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology. 2012;56:434-443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 80. | Fernholz D, Stemler M, Brunetto M, Bonino F, Will H. Replicating and virion secreting hepatitis B mutant virus unable to produce preS2 protein. J Hepatol. 1991;13 Suppl 4:S102-S104. [PubMed] [Cited in This Article: ] |
| 81. | Gerken G, Kremsdorf D, Capel F, Petit MA, Dauguet C, Manns MP, Meyer zum Büschenfelde KH, Brechot C. Hepatitis B defective virus with rearrangements in the preS gene during chronic HBV infection. Virology. 1991;183:555-565. [PubMed] [Cited in This Article: ] |
| 82. | Sugauchi F, Ohno T, Orito E, Sakugawa H, Ichida T, Komatsu M, Kuramitsu T, Ueda R, Miyakawa Y, Mizokami M. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003;70:537-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 94] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 83. | Chen CH, Changchien CS, Lee CM, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis. 2008;198:1634-1642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | Cao L, Wu C, Shi H, Gong Z, Zhang E, Wang H, Zhao K, Liu S, Li S, Gao X. Coexistence of hepatitis B virus quasispecies enhances viral replication and the ability to induce host antibody and cellular immune responses. J Virol. 2014;88:8656-8666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 85. | Summers J, Smith PM, Horwich AL. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819-2824. [PubMed] [Cited in This Article: ] |
| 86. | Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565-4571. [PubMed] [Cited in This Article: ] |
| 87. | Lenhoff RJ, Luscombe CA, Summers J. Acute liver injury following infection with a cytopathic strain of duck hepatitis B virus. Hepatology. 1999;29:563-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 88. | Summers J, Smith PM, Huang MJ, Yu MS. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310-1317. [PubMed] [Cited in This Article: ] |
| 89. | Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol. 2007;81:6164-6174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 90. | Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007;81:12472-12484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 234] [Cited by in F6Publishing: 255] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 91. | Lentz TB, Loeb DD. Roles of the envelope proteins in the amplification of covalently closed circular DNA and completion of synthesis of the plus-strand DNA in hepatitis B virus. J Virol. 2011;85:11916-11927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 92. | Mu SC, Lin YM, Jow GM, Chen BF. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J Hepatol. 2009;50:264-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 93. | Chirara MM, Chetsanga CJ. Variant of hepatitis B virus isolated in Zimbabwe. J Med Virol. 1994;42:73-78. [PubMed] [Cited in This Article: ] |
| 94. | Bowyer SM, van Staden L, Kew MC, Sim JG. A unique segment of the hepatitis B virus group A genotype identified in isolates from South Africa. J Gen Virol. 1997;78:1719-1729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 135] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 95. | Owiredu WK, Kramvis A, Kew MC. Molecular analysis of hepatitis B virus genomes isolated from black African patients with fulminant hepatitis B. J Med Virol. 2001;65:485-492. [PubMed] [Cited in This Article: ] |
| 96. | Garfein RS, Bower WA, Loney CM, Hutin YJ, Xia GL, Jawanda J, Groom AV, Nainan OV, Murphy JS, Bell BP. Factors associated with fulminant liver failure during an outbreak among injection drug users with acute hepatitis B. Hepatology. 2004;40:865-873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 97. | Liaw YF, Leung N, Kao JH, Piratvisuth T, Gane E, Han KH, Guan R, Lau GK, Locarnini S; Chronic Hepatitis B Guideline Working Party of the Asian-Pacific Association for the Study of the Liver. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2008 update. Hepatol Int. 2008;2:263-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 666] [Cited by in F6Publishing: 724] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 98. | Shen T, Yan XM, Zou YL, Gao JM, Dong H. Virologic characteristics of hepatitis B virus in patients infected via maternal-fetal transmission. World J Gastroenterol. 2008;14:5674-5682. [PubMed] [Cited in This Article: ] |
| 99. | Shen FC, Su IJ, Wu HC, Hsieh YH, Yao WJ, Young KC, Chang TC, Hsieh HC, Tsai HN, Huang W. A pre-S gene chip to detect pre-S deletions in hepatitis B virus large surface antigen as a predictive marker for hepatoma risk in chronic hepatitis B virus carriers. J Biomed Sci. 2009;16:84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 100. | Shen T, Yan XM, Zhang JP, Wang JL, Zuo RX, Li L, Wang LP. Evolution of Hepatitis B Virus in a Chronic HBV-Infected Patient over 2 Years. Hepat Res Treat. 2011;2011:939148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 101. | Heo NY, Lee HC, Park YK, Park JW, Lim YS, Kim KM, Shim JH, Lee YJ. Lack of association between hepatitis B virus pre-S mutations and recurrence after surgical resection in hepatocellular carcinoma. J Med Virol. 2013;85:589-596. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 102. | Hung CH, Chen CH, Lee CM, Hu TH, Lu SN, Wang JH, Huang CM. Role of viral genotypes and hepatitis B viral mutants in the risk of hepatocellular carcinoma associated with hepatitis B and C dual infection. Intervirology. 2013;56:316-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 103. | Fan YF, Lu CC, Chen WC, Yao WJ, Wang HC, Chang TT, Lei HY, Shiau AL, Su IJ. Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology. 2001;33:277-286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 155] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 104. | Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007;133:1466-1474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 197] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 105. | Ghosh S, Mondal RK, Banerjee P, Nandi M, Sarkar S, Das K, Santra A, Banerjee S, Chowdhury A, Datta S. Tracking the naturally occurring mutations across the full-length genome of hepatitis B virus of genotype D in different phases of chronic e-antigen-negative infection. Clin Microbiol Infect. 2012;18:E412-E418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 106. | Xu Z, Yen TS. Intracellular retention of surface protein by a hepatitis B virus mutant that releases virion particles. J Virol. 1996;70:133-140. [PubMed] [Cited in This Article: ] |
| 107. | Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, Huang W. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 108. | Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77-100. [PubMed] [Cited in This Article: ] |
| 109. | Hildt E, Hofschneider PH. The PreS2 activators of the hepatitis B virus: activators of tumour promoter pathways. Recent Results Cancer Res. 1998;154:315-329. [PubMed] [Cited in This Article: ] |
| 110. | Caselmann WH, Meyer M, Kekulé AS, Lauer U, Hofschneider PH, Koshy R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci USA. 1990;87:2970-2974. [PubMed] [Cited in This Article: ] |
| 111. | Lauer U, Weiss L, Lipp M, Hofschneider PH, Kekulé AS. The hepatitis B virus preS2/St transactivator utilizes AP-1 and other transcription factors for transactivation. Hepatology. 1994;19:23-31. [PubMed] [Cited in This Article: ] |
| 112. | Sterneck M, Günther S, Gerlach J, Naoumov NV, Santantonio T, Fischer L, Rogiers X, Greten H, Williams R, Will H. Hepatitis B virus sequence changes evolving in liver transplant recipients with fulminant hepatitis. J Hepatol. 1997;26:754-764. [PubMed] [Cited in This Article: ] |
| 113. | Pollicino T, Zanetti AR, Cacciola I, Petit MA, Smedile A, Campo S, Sagliocca L, Pasquali M, Tanzi E, Longo G. Pre-S2 defective hepatitis B virus infection in patients with fulminant hepatitis. Hepatology. 1997;26:495-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 83] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 114. | Bock CT, Tillmann HL, Maschek HJ, Manns MP, Trautwein C. A preS mutation isolated from a patient with chronic hepatitis B infection leads to virus retention and misassembly. Gastroenterology. 1997;113:1976-1982. [PubMed] [Cited in This Article: ] |
| 115. | Bock CT, Kubicka S, Manns MP, Trautwein C. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology. 1999;29:1236-1247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 55] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 116. | Bock CT, Tillmann HL, Manns MP, Trautwein C. The pre-S region determines the intracellular localization and appearance of hepatitis B virus. Hepatology. 1999;30:517-525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 47] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 117. | Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220-9231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1361] [Cited by in F6Publishing: 1428] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 118. | Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576-1582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 564] [Cited by in F6Publishing: 573] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 119. | Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 673] [Cited by in F6Publishing: 733] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 120. | Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460-1471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 344] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 121. | Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239-247. [PubMed] [Cited in This Article: ] |
| 122. | Lépine S, Allegood JC, Park M, Dent P, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 2011;18:350-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 123. | Sakaki K, Wu J, Kaufman RJ. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem. 2008;283:15370-15380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 156] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 124. | Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 2007;3:160-162. [PubMed] [Cited in This Article: ] |
| 125. | Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299-30304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 712] [Cited by in F6Publishing: 755] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 126. | Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383-4388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 127. | Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319-6333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 128. | Naoumov NV, Eddleston AL. Host immune response and variations in the virus genome: pathogenesis of liver damage caused by hepatitis B virus. Gut. 1994;35:1013-1017. [PubMed] [Cited in This Article: ] |
| 129. | Bertoletti A, Ferrari C. Adaptive immunity in HBV infection. J Hepatol. 2016;64:S71-S83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 279] [Cited by in F6Publishing: 300] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 130. | Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 555] [Cited by in F6Publishing: 557] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 131. | Peeridogaheh H, Meshkat Z, Habibzadeh S, Arzanlou M, Shahi JM, Rostami S, Gerayli S, Teimourpour R. Current concepts on immunopathogenesis of hepatitis B virus infection. Virus Res. 2018;245:29-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 132. | Ando K, Moriyama T, Guidotti LG, Wirth S, Schreiber RD, Schlicht HJ, Huang SN, Chisari FV. Mechanisms of class I restricted immunopathology. A transgenic mouse model of fulminant hepatitis. J Exp Med. 1993;178:1541-1554. [PubMed] [Cited in This Article: ] |
| 133. | Mina T, Amini Bavil Olyaee S, Tacke F, Maes P, Van Ranst M, Pourkarim MR. Genomic Diversity of Hepatitis B Virus Infection Associated With Fulminant Hepatitis B Development. Hepat Mon. 2015;15:e29477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 134. | Bertoletti A, Maini M, Williams R. Role of hepatitis B virus specific cytotoxic T cells in liver damage and viral control. Antiviral Res. 2003;60:61-66. [PubMed] [Cited in This Article: ] |
| 135. | Maini MK, Boni C, Lee CK, Larrubia JR, Reignat S, Ogg GS, King AS, Herberg J, Gilson R, Alisa A. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J Exp Med. 2000;191:1269-1280. [PubMed] [Cited in This Article: ] |
| 136. | Bertoletti A, Maini MK. Protection or damage: a dual role for the virus-specific cytotoxic T lymphocyte response in hepatitis B and C infection? Curr Opin Immunol. 2000;12:403-408. [PubMed] [Cited in This Article: ] |
| 137. | Bagaglio S, Albarello L, Biswas P, Uberti-Foppa C, Fortis C, Morsica G. Virological pattern of hepatitis B infection in an HIV-positive man with fatal fulminant hepatitis B: a case report. J Med Case Rep. 2009;3:110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 138. | Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxì A, Donato F, Ferrari C, Gaeta GB. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652-657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 562] [Cited by in F6Publishing: 574] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 139. | Makvandi M. Update on occult hepatitis B virus infection. World J Gastroenterol. 2016;22:8720-8734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 111] [Cited by in F6Publishing: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 140. | Chemin I, Trépo C. Clinical impact of occult HBV infections. J Clin Virol. 2005;34 Suppl 1:S15-S21. [PubMed] [Cited in This Article: ] |
| 141. | Hashemi SJ, Hajiani E, Masjedizadeh A, Makvandi M, Shayesteh AA, Alavinejad SP, Kadkhodaei A, Shahbazian H, Jasemi F, Karimi M. Occult hepatitis B infection in patients with cryptogenic liver cirrhosis in southwest of Iran. Jundishapur J Microbiol. 2015;8:e16873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 142. | Saitta C, Tripodi G, Barbera A, Bertuccio A, Smedile A, Ciancio A, Raffa G, Sangiovanni A, Navarra G, Raimondo G. Hepatitis B virus (HBV) DNA integration in patients with occult HBV infection and hepatocellular carcinoma. Liver Int. 2015;35:2311-2317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 143. | Chemin I, Zoulim F, Merle P, Arkhis A, Chevallier M, Kay A, Cova L, Chevallier P, Mandrand B, Trépo C. High incidence of hepatitis B infections among chronic hepatitis cases of unknown aetiology. J Hepatol. 2001;34:447-454. [PubMed] [Cited in This Article: ] |
| 144. | Pollicino T, Squadrito G, Cerenzia G, Cacciola I, Raffa G, Craxi A, Farinati F, Missale G, Smedile A, Tiribelli C. Hepatitis B virus maintains its pro-oncogenic properties in the case of occult HBV infection. Gastroenterology. 2004;126:102-110. [PubMed] [Cited in This Article: ] |
| 145. | Wong DK, Huang FY, Lai CL, Poon RT, Seto WK, Fung J, Hung IF, Yuen MF. Occult hepatitis B infection and HBV replicative activity in patients with cryptogenic cause of hepatocellular carcinoma. Hepatology. 2011;54:829-836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 113] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 146. | Altfeld M, Rockstroh JK, Addo M, Kupfer B, Pult I, Will H, Spengler U. Reactivation of hepatitis B in a long-term anti-HBs-positive patient with AIDS following lamivudine withdrawal. J Hepatol. 1998;29:306-309. [PubMed] [Cited in This Article: ] |
| 147. | Kidd-Ljunggren K, Simonsen O. Reappearance of hepatitis B 10 years after kidney transplantation. N Engl J Med. 1999;341:127-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 148. | Westhoff TH, Jochimsen F, Schmittel A, Stoffler-Meilicke M, Schafer JH, Zidek W, Gerlich WH, Thiel E. Fatal hepatitis B virus reactivation by an escape mutant following rituximab therapy. Blood. 2003;102:1930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 178] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 149. | Kubo S, Tamori A, Ohba K, Shuto T, Yamamoto T, Tanaka H, Nishiguchi S, Wakasa K, Hirohashi K, Kinoshita H. Previous or occult hepatitis B virus infection in hepatitis C virus-associated hepatocellular carcinoma without hepatic fibrosis. Dig Dis Sci. 2001;46:2408-2414. [PubMed] [Cited in This Article: ] |
| 150. | Lalazar G, Rund D, Shouval D. Screening, prevention and treatment of viral hepatitis B reactivation in patients with haematological malignancies. Br J Haematol. 2007;136:699-712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 238] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 151. | García-Fulgueiras A, García-Pina R, Morant C, García-Ortuzar V, Génova R, Alvarez E. Hepatitis C and hepatitis B-related mortality in Spain. Eur J Gastroenterol Hepatol. 2009;21:895-901. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 152. | Marcellin P, Pequignot F, Delarocque-Astagneau E, Zarski JP, Ganne N, Hillon P, Antona D, Bovet M, Mechain M, Asselah T. Mortality related to chronic hepatitis B and chronic hepatitis C in France: evidence for the role of HIV coinfection and alcohol consumption. J Hepatol. 2008;48:200-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 153. | Xie M, Rao W, Yang T, Deng Y, Zheng H, Pan C, Liu Y, Shen Z, Jia J. Occult hepatitis B virus infection predicts de novo hepatitis B infection in patients with alcoholic cirrhosis after liver transplantation. Liver Int. 2015;35:897-904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 154. | Candotti D, Grabarczyk P, Ghiazza P, Roig R, Casamitjana N, Iudicone P, Schmidt M, Bird A, Crookes R, Brojer E. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J Hepatol. 2008;49:537-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 155. | Allain JP, Belkhiri D, Vermeulen M, Crookes R, Cable R, Amiri A, Reddy R, Bird A, Candotti D. Characterization of occult hepatitis B virus strains in South African blood donors. Hepatology. 2009;49:1868-1876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 156. | Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, Yeh SH, Yu H, Xue Y, Chen YX. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J Hepatol. 2012;57:720-729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 157. | Saha D, Pal A, Sarkar N, Das D, Blackard JT, Guha SK, Saha B, Chakravarty R. Occult hepatitis B virus infection in HIV positive patients at a tertiary healthcare unit in eastern India. PLoS One. 2017;12:e0179035. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 158. | Ye Q, Shang SQ, Li W. A new vaccine escape mutant of hepatitis B virus causes occult infection. Hum Vaccin Immunother. 2015;11:407-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 159. | Hou J, Wang Z, Cheng J, Lin Y, Lau GK, Sun J, Zhou F, Waters J, Karayiannis P, Luo K. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology. 2001;34:1027-1034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 157] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 160. | Kwei K, Tang X, Lok AS, Sureau C, Garcia T, Li J, Wands J, Tong S. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J Virol. 2013;87:2352-2357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 161. | Biswas S, Candotti D, Allain JP. Specific amino acid substitutions in the S protein prevent its excretion in vitro and may contribute to occult hepatitis B virus infection. J Virol. 2013;87:7882-7892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 162. | Zuckerman JN, Zuckerman AJ. Mutations of the surface protein of hepatitis B virus. Antiviral Res. 2003;60:75-78. [PubMed] [Cited in This Article: ] |
| 163. | Xiang KH, Michailidis E, Ding H, Peng YQ, Su MZ, Li Y, Liu XE, Dao Thi VL, Wu XF, Schneider WM. Effects of amino acid substitutions in hepatitis B virus surface protein on virion secretion, antigenicity, HBsAg and viral DNA. J Hepatol. 2017;66:288-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 164. | Waters JA, Kennedy M, Voet P, Hauser P, Petre J, Carman W, Thomas HC. Loss of the common “A” determinant of hepatitis B surface antigen by a vaccine-induced escape mutant. J Clin Invest. 1992;90:2543-2547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 154] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 165. | Shahmoradi S, Yahyapour Y, Mahmoodi M, Alavian SM, Fazeli Z, Jazayeri SM. High prevalence of occult hepatitis B virus infection in children born to HBsAg-positive mothers despite prophylaxis with hepatitis B vaccination and HBIG. J Hepatol. 2012;57:515-521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 166. | Jeantet D, Chemin I, Mandrand B, Tran A, Zoulim F, Merle P, Trepo C, Kay A. Cloning and expression of surface antigens from occult chronic hepatitis B virus infections and their recognition by commercial detection assays. J Med Virol. 2004;73:508-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 167. | Minuk GY, Sun DF, Uhanova J, Zhang M, Caouette S, Nicolle LE, Gutkin A, Doucette K, Martin B, Giulivi A. Occult hepatitis B virus infection in a North American community-based population. J Hepatol. 2005;42:480-485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 168. | Wands JR, Marciniak RA, Isselbacher KJ, Varghese M, Don G, Halliday JW, Powell LW. Demonstration of previously undetected hepatitis B viral determinants in an Australian Aboriginal population by monoclonal anti-hbs antibody radioimmunoassays. Lancet. 1982;1:977-980. [PubMed] [Cited in This Article: ] |
| 169. | Weinberger KM, Bauer T, Böhm S, Jilg W. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. J Gen Virol. 2000;81:1165-1174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 165] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 170. | Zaaijer HL, Torres P, Ontañón A, Ponte LG, Koppelman MH, Lelie PN, Hemert FJ, Boot HJ. Multiple surface antigen mutations in five blood donors with occult hepatitis B virus infection. J Med Virol. 2008;80:1344-1349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 171. | Gish RG, Yuen MF, Chan HL, Given BD, Lai CL, Locarnini SA, Lau JY, Wooddell CI, Schluep T, Lewis DL. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015;121:97-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 172. | Schluep T, Lickliter J, Hamilton J, Lewis DL, Lai CL, Lau JY, Locarnini SA, Gish RG, Given BD. Safety, Tolerability, and Pharmacokinetics of ARC-520 Injection, an RNA Interference-Based Therapeutic for the Treatment of Chronic Hepatitis B Virus Infection, in Healthy Volunteers. Clin Pharmacol Drug Dev. 2017;6:350-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 173. | Zobeiri M. Occult hepatitis B: clinical viewpoint and management. Hepat Res Treat. 2013;2013:259148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |