Published online Jan 14, 2015. doi: 10.3748/wjg.v21.i2.699
Peer-review started: April 20, 2014
First decision: May 13, 2014
Revised: June 10, 2014
Accepted: July 11, 2014
Article in press: July 11, 2014
Published online: January 14, 2015
Processing time: 273 Days and 5.2 Hours
Genotyping is conclusive for the diagnosis of progressive familial intrahepatic cholestasis type 3 (PFIC3). Here we report a Chinese patient of PFIC3 with compound mutations in the ABCB4 gene. Liver biopsy was performed on a 17-year-old male patient with intrahepatic cholestasis of unknown etiology. Liver histology findings are indicative of intrahepatic cholestasis with extensive fibrosis. Genotyping revealed c.175C>T (p.L59L) mutation in exon 4, c.504C>T (p.N168N) mutation in exon 6, c.711A>T (p.I237I) mutation in exon 8, c.874A>T (p.K292X) in exon 9 and a novel mutation, c.1804G>T (p.G602W) in exon 15. Based on these findings, the patient was diagnosed with PFIC3. The novel mutation p.G602W in exon 15 was predicted as probably damaging by PolyPhen-2 with a score of 0.986 (sensitivity: 0.54; specificity: 0.94) and was predicted to affect protein function with a SIFT score of 0.01.
Core tip: This study described a 17-year-old Chinese male patient with a 2 years history of intrahepatic cholestasis of unknown etiology who was later diagnosed with progressive familial intrahepatic cholestasis type 3 through clinical findings and gene analysis which revealed multiple mutations in the ABCB4 gene. One novel mutation of the ABCB4 gene p.G602W has also been identified. The novel mutation p.G602W in exon 15 was predicted as probably damaging by PolyPhen-2, with a score of 0.986, and was predicted to affect protein function with a SIFT score of 0.01.
- Citation: Sun HZ, Shi H, Zhang SC, Shen XZ. Novel mutation in a Chinese patient with progressive familial intrahepatic cholestasis type 3. World J Gastroenterol 2015; 21(2): 699-703
- URL: https://www.wjgnet.com/1007-9327/full/v21/i2/699.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i2.699
Progressive familial intrahepatic cholestasis (PFIC) refers to a heterogeneous group of autosomal recessive liver disorders in childhood that disrupt bile formation and present with cholestasis of hepatocellular origin[1]. There are three types of PFIC, each caused by mutations in different hepatocellular transport system genes involved in bile formation. PFIC type 1 (PFIC1) is due to mutations in the ATP8B1 gene; PFIC type 2 (PFIC2) is due to mutations in the ABCB11 gene; and PFIC type 3 (PFIC3) is due to mutations in the ABCB4 gene [also designated multidrug resistant 3 (MDR3)][2]. The non-specific clinical signs and pathological findings of this disease make the diagnosis of PFIC difficult without the use of gene analysis.
In the present report, we describe a 17-year-old Chinese male patient with a 2 years history of intrahepatic cholestasis of unknown etiology who was later diagnosed with PFIC3 through clinical findings and gene analysis which revealed multiple mutations in the ABCB4 gene consistent with the disease.
A 17-year-old Chinese male presented with a history of pruritus and abnormal liver function for 2 years with unknown etiology by the time of admission to our hospital. No fever, weight loss, abdominal pain, rash, or joint pain was documented. The patient denies a history of hepatitis or use of any hepatotoxic drugs. He does not smoke, consume alcohol and denies any family history of hereditary or infectious disease.
On physical examination, the patient was notable for jaundice. No palmar erythema or spider angioma was noted. There was no enlargement of his superficial lymph nodes. His respiratory system and cardiac examination were normal. Abdominal examination revealed hepatosplenomegaly. Shifting dullness was absent. Examination of the musculoskeletal system was within normal limits. Lower extremity edema was negative.
Results of laboratory tests done during the time of admission were as follows: total bilirubin: 63.6 μmol/L, conjugated bilirubin: 47.9 μmol/L, total protein: 61 g/L, albumin: 34 g/L, alanine aminotransferase: 330 U/L, aspartate aminotransferase: 220 U/L, alkaline phosphatase: 495 U/L, gamma-glutamyltransferase (GGT): 388 U/L, total bile acid: 198.2 μmol/L, lactate dehydrogenase: 250 U/L. Hepatitis A, hepatitis B, hepatitis C and hepatitis E viral markers: negative. An autoantibody panel (including antinuclear antibodies, anti-smooth muscle antibodies, anti-mitochondrial antibody, anti-mitochondrial antibody M2 subtype, anti-centromere antibody, anti-histone antibodies, anti-JO-1 antibody, anti-nucleosome antibodies, anti-ribosomal P protein antibodies, anti-proliferating cell nuclear antigen (anti-PCNA) antibody, anti-polymyositis-scleroderma (anti-PM-Scl) antibody, anti-ribonucleo protein (anti-RNP) antibodies, anti-topoisomerase I (anti-SCL-70) antibodies, anti-Sm antibodies, anti-Sjogren’s syndrome antigen A (anti-SS-A) antibody, anti-Sjogren’s syndrome antigen B (anti-SS-B) antibody, neutrophil cytoplasmic antibody, anti-double-stranded DNA antibodies, anti-glomerular basement membrane antibodies, anti-kidney microsomal antibody type I, anti-soluble liver/liver pancreas antigen-antibody) was negative. The ceruloplasmin and tumor marker panels were within normal limits. Abdominal magnetic resonance imaging and magnetic resonance cholangiopancreatography revealed nodules in segment VI and segment VIII of the liver, highly suspicious of focal nodular hyperplasia. There was also evidence of hepatosplenomegaly and portal hypertension. Liver histology obtained from a biopsy exhibited disorganized lobular structure and evidence of cholestasis, marked dilation of bile canaliculi and in some ductules containing bile plugs. Signs of chronic inflammation and fibrosis were found in the portal area. Immunohistochemical analysis with cytokeratin (CK) 7 and CK19 revealed ductular proliferation in the portal area. Reticular stain showed extensive portal fibrosis and the formation of pseudolobules. These findings are indicative of intrahepatic cholestasis with extensive fibrosis (stage 3). Genomic DNA was purified from peripheral blood. All of the 27 coding exons of ABCB4 together with at least 100 bp of the adjacent intronic sequence were amplified by polymerase chain reaction and directly sequenced. Based on the results of ABCB4 gene analysis (Table 1), a diagnosis of PFIC3 was made.
| Exon | Result | SNP No. |
| Exon 4 | c.175 C>T (p.L59L) (heterozygous) | rs2302387 |
| Exon 6 | c.504 C>T (p.N168N) (heterozygous) | rs1202283 |
| Exon 8 | c.711 A>T (p.I237I) (heterozygous) | rs2109505 |
| Exon 9 | c.874 A>T (p.K292X) (heterozygous) | CM072813 |
| Exon 15 | c.1804 G>T (p.G602W) (heterozygous) |
In this study, we present a 17-year-old Chinese male patient with a novel mutation in ABCB4, who experienced a two years history of intrahepatic cholestasis of unknown etiology. Liver histology findings were indicative of intrahepatic cholestasis with extensive fibrosis (stage 3). We excluded other causes of cholestasis such as primary biliary cirrhosis, sclerosing cholangitis, drug induced liver injury, alpha-1 antitrypsin deficiency, cystic fibrosis, etc. At this point, there was a high index of suspicion of PFIC. Patients with PFIC1 and PFIC2 have normal serum GGT activity, while patients with PFIC3 have high serum GGT activity. PFIC3 patients can also be distinguished from patients with PFIC types 1-2 in that they rarely present with cholestatic jaundice in the neonatal period, but do so later on in infancy, childhood or young adulthood instead[3]. The patient’s clinical features (present symptomatically at a later age), biochemical results (markedly elevated GGT) and histological findings (presence of ductular proliferation) indicated the suspected diagnosis of PFIC3 and lead to the genetic analysis of the ABCB4 gene accordingly. Liver immunostaining of MDR3 was not considered in this patient because patients with ABCB4 mutations exhibit variable MDR3 canalicular immunostaining[4]. Normal liver immunostaining does not preclude a gene defect, as a mutation may induce loss of function, but indicates normal synthesis and expression[3]. Furthermore, it is reported that MDR3 immunostaining on frozen liver biopsy samples is not a sensitive diagnostic tool for the detection of heterozygous MDR3/ABCB4 gene mutations[5]. Results of the gene analysis showed that the patient is a compound heterozygote for multiple mutations reported in literature predicted to impair MDR3 function. Combined with his clinical features and laboratory findings, the diagnosis of PFIC3 was reached.
PFIC3 is caused by a genetic defect in the ABCB4 gene located on chromosome 7[3]. MDR3 in humans are phospholipid translocators involved in biliary phosphatidylcholine excretion and are predominantly, if not exclusively, expressed in the canalicular membrane of the hepatocyte[4]. Cholestasis results from the toxicity of bile in which detergent bile salts are not inactivated by phospholipids, leading to bile canaliculi and biliary epithelium injuries. The mechanism of liver damage in PFIC3 patients is most likely related to the absence of biliary phospholipids. Injury to bile canaliculi and biliary epithelium is probably due to continuous exposure to hydrophobic bile salts, the detergent effects of which are no longer being countered by phospholipids, thereby leading to cholangitis. In addition, the stability of mixed micelles in bile is determined by a three-phase system in which the proper proportion of bile salts and phospholipids is necessary for maintaining solubility of cholesterol. The absence of phospholipids in bile would be expected to destabilize micelles and promote bile lithogenicity through crystallization of cholesterol, which might favor small bile duct obstruction[3].
According to the Human Genome Mutation Database (http://www.hgmd.org/)[6], there are over 150 disease associated mutations in the ABCB4 gene identified, 50 of them associated with PFIC3. These include missense mutations, nonsense mutations, splice site mutations, insertions and deletions. Among them, patients with nonsense mutations and deletions of the ABCB4 gene are associated with a more severe clinical course of the disease[7].
As reported in the sequence annotations of Uniprot database for ABCB4, accession P21439, the domains essential for organic molecule efflux are arranged along the ABCB4 protein sequence in the amino acid ranges 57-359 (1st ABC transmembrane transporter), 394-630 (1st ABC transporter), 711-999 (2nd ABC transmembrane transporter) and 1034-1279 (2nd ABC transporter). In our patient, three heterozygous silent mutations p.L59L, p.N168N and p.I237I were observed, all of which have been previously reported[8,9]. Additionally, one heterozygous nonsense mutation p.K292X and a novel heterozygous missense mutation pG602W were also found. Location of the mutations relative to their location to the respective domains in the protein sequence can be found in Figure 1.
In some patients with a PFIC3 phenotype, only one mutated allele or no mutation is identified[4]. This situation may be explained by mutations that map to the regulatory sequences of the gene. It is also possible that other unidentified genes involved in bile formation may be responsible for the PFIC1-3 phenotypes[2,3]. Modifier genes and environmental influences could also play a role in the expression of PFIC[3]. Another interesting possibility are heterozygous statuses, where a mutated protein may have a dominant negative effect on MDR3 expression or function[4].
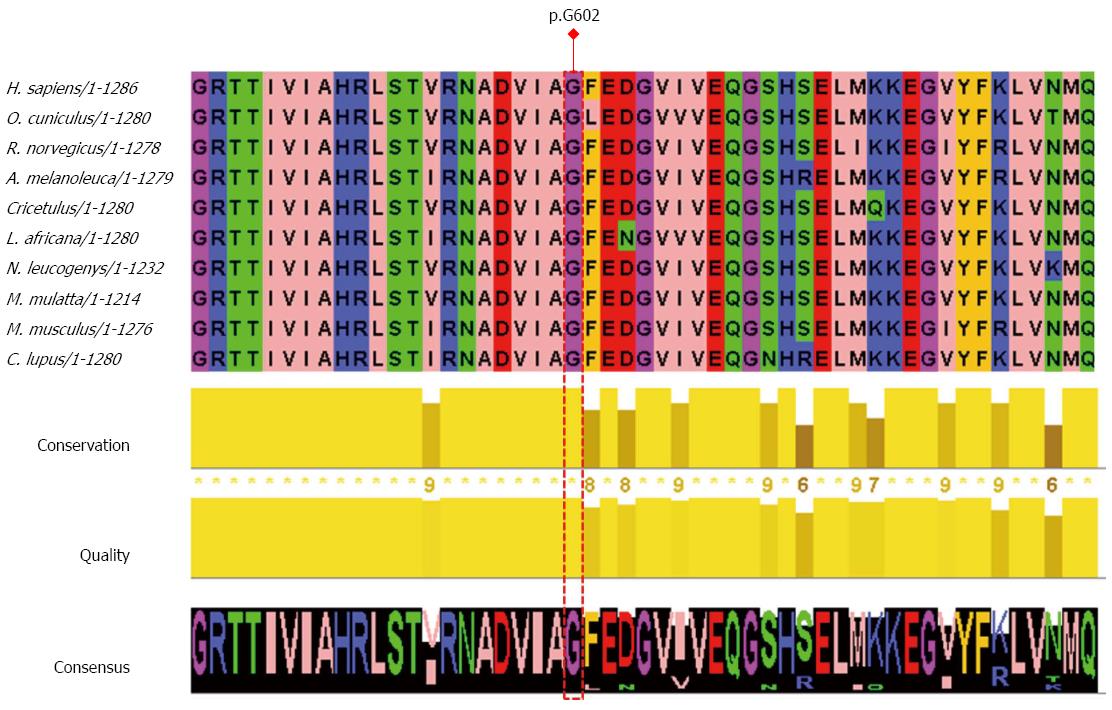
To evaluate the functional significance of the novel mutation p.G602W, PolyPhen-2 (http://genetics.bwh.harvard.edu/pph/) and SIFT (http://sift.jcvi.org/) were used. PolyPhen-2 uses the Naïve Bayes classifier, trained using supervised machine-learning to predict the impact of an allele replacement from its individual features and mutations are appraised qualitatively, as benign, possibly damaging, or probably damaging[10]. Using the HumVar model which distinguishes mutations with drastic effects from all the remaining human variations with a score range between 0 to 1 (values close to 0 are benign, whereas those near 1 are probably damaging), the novel mutation p.G602W was predicted as probably damaging by PolyPhen-2 with a score of 0.986 (sensitivity: 0.54; specificity: 0.94). Furthermore, multiple sequence alignment of sequences, derived from PolyPhen-2 and edited with the help of Jalview[11], showed that the novel mutation p.G602W is at a location which is physicochemically conserved in most mammals (Figure 2). Among the sequences aligned, none of them included a tryptophan at the position corresponding to glycine 602 of MDR3. All other sequences mostly harbored a glycine at the corresponding position depicting a highly conserved amino acid.
SIFT is a sequence homology-based tool that sorts intolerant from tolerant amino acid substitutions and predicts whether an amino acid substitution in a protein will have a phenotypic effect. SIFT is based on the premise that protein evolution is correlated with protein function. Positions important for function should be conserved in an alignment of the protein family, whereas unimportant positions should appear diverse in an alignment. SIFT predicts substitutions with scores less than 0.05 as deleterious[12]. Results from SIFT showed that the novel mutation p.G602W was predicted to affect protein function with a SIFT score of 0.01.
The patient does not have any family history and neither parent is symptomatic. With regard to clinical features, a family history for diseases associated with ABCB4 deficiency was observed in two thirds of patients harboring ABCB4 mutations. Patients with mild genotypes, including single heterozygous mutations, have variable expression of liver disease that may be influenced by comorbidity or triggering factors and modulated by still unknown genetic modifiers[9].
In conclusion, a diagnosis of PFIC should be suspected in patients with a clinical history of cholestasis of unknown origin after exclusion of other common causes of cholestasis. Genetic analysis remains an important tool for a conclusive diagnosis of PFIC3 when combined with clinical, biochemical, radiological and histological studies. From our in silico analysis of the novel mutation p.G602W in exon 15, the highly conserved glycine 602 of human MDR3 is predicted to be essential for proper MDR3 function. This finding will be useful in future studies to further elucidate the pathogenic mechanisms of ABCB4 mutations at the molecular level.
A 17-year-old Chinese male presented with a history of pruritus and jaundice for 2 years.
Abdominal examination revealed hepatosplenomegaly.
The authors excluded other causes of cholestasis such as primary biliary cirrhosis, sclerosing cholangitis, drug induced liver injury, alpha-1 antitrypsin deficiency, cystic fibrosis, etc.
Total bilirubin: 63.6 μmol/L, conjugated bilirubin: 47.9 μmol/L, total protein: 61 g/L, albumin: 34 g/L, alanine aminotransferase: 330 U/L, aspartate aminotransferase: 220 U/L, alkaline phosphatase: 495 U/L, gamma-glutamyltransferase: 388 U/L, total bile acid: 198.2 μmol/L, lactate dehydrogenase: 250 U/L. hepatitis viral markers and autoantibody panel were all negative.
Abdominal magnetic resonance imaging and magnetic resonance cholangiopancreatography revealed focal nodular hyperplasia in segment VI and segment VIII of the liver, hepatosplenomegaly and portal hypertension.
Liver histology obtained from biopsy revealed ductular proliferation and portal fibrosis.
The patient was treated with ursodeoxycholic acid.
This case report a novel mutation in a Chinese patient with progressive familial intrahepatic cholestasis type 3 (PFIC3); the novel mutation p.G602W in exon 15 was predicted as probably damaging by PolyPhen-2 with a score of 0.986 and was predicted to affect protein function with a SIFT score of 0.01.
The discovery and recording of a novel mutation in a Chinese patient with PFIC3 is of high clinical importance and can contribute to the recording of mutations.
P- Reviewer: Chan ACY, Shike H, Vasilieva LE, Wang JS S- Editor: Nan J L- Editor: O’Neill M E- Editor: Ma S
| 1. | Hori T, Nguyen JH, Uemoto S. Progressive familial intrahepatic cholestasis. Hepatobiliary Pancreat Dis Int. 2010;9:570-578. [PubMed] |
| 2. | Davit-Spraul A, Gonzales E, Baussan C, Jacquemin E. Progressive familial intrahepatic cholestasis. Orphanet J Rare Dis. 2009;4:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 244] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 3. | Jacquemin E. Progressive familial intrahepatic cholestasis. Clin Res Hepatol Gastroenterol. 2012;36 Suppl 1:S26-S35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 4. | Davit-Spraul A, Gonzales E, Baussan C, Jacquemin E. The spectrum of liver diseases related to ABCB4 gene mutations: pathophysiology and clinical aspects. Semin Liver Dis. 2010;30:134-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 5. | Sannier A, Ganne N, Tepper M, Ziol M. MDR3 immunostaining on frozen liver biopsy samples is not a sensitive diagnostic tool for the detection of heterozygous MDR3/ABCB4 gene mutations. Virchows Arch. 2012;460:535-537; author reply 539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1056] [Cited by in RCA: 1044] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 7. | Fang LJ, Wang JS. [Progressive familial intrahepatic cholestasis type 3]. Zhonghua Gan Zang Bing Za Zhi. 2010;18:238-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Lang T, Haberl M, Jung D, Drescher A, Schlagenhaufer R, Keil A, Mornhinweg E, Stieger B, Kullak-Ublick GA, Kerb R. Genetic variability, haplotype structures, and ethnic diversity of hepatic transporters MDR3 (ABCB4) and bile salt export pump (ABCB11). Drug Metab Dispos. 2006;34:1582-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Colombo C, Vajro P, Degiorgio D, Coviello DA, Costantino L, Tornillo L, Motta V, Consonni D, Maggiore G. Clinical features and genotype-phenotype correlations in children with progressive familial intrahepatic cholestasis type 3 related to ABCB4 mutations. J Pediatr Gastroenterol Nutr. 2011;52:73-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10971] [Cited by in RCA: 10411] [Article Influence: 694.1] [Reference Citation Analysis (0)] |
| 11. | Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189-1191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8558] [Cited by in RCA: 7218] [Article Influence: 451.1] [Reference Citation Analysis (0)] |
| 12. | Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812-3814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4033] [Cited by in RCA: 4463] [Article Influence: 202.9] [Reference Citation Analysis (0)] |