Published online Dec 14, 2007. doi: 10.3748/wjg.v13.i46.6243
Revised: August 27, 2007
Accepted: September 20, 2007
Published online: December 14, 2007
AIM: To investigate the safety of β-L-D4A on DNA polymerase α.
METHODS: Ion exchange chromatography was used to separate DNA polymerase α from crude extract of human Hela cells. Detailed kinetic parameters were determined for β-L-D4A against DNA polymerase α.
RESULTS: DNA polymerase α was purified with 4% yield and 31 000 units/mg specific activity. The Michaelis constant (Km = 3.22 μmol/L), 50% inhibition concentration (IC50 = 178.49 μmol/L) and inhibition constant (Ki = 126 μmol/L) of β-L-D4A were determined by kinetic analysis.
CONCLUSION: β-L-D4A is a more safe nucleoside for hepatitis B virus (HBV) infection with a lower host toxicity.
- Citation: Li Y, Lin JS, Zhang YH, Wang XY, Chang Y, He XX. Effect and mechanism of β-L-D4A on DNA polymerase α. World J Gastroenterol 2007; 13(46): 6243-6248
- URL: https://www.wjgnet.com/1007-9327/full/v13/i46/6243.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i46.6243
Infection with hepatitis B virus (HBV) is a global medical problem[1]. With the support of National Natural Science Foundation of China[2], we have found a novel nucleoside analog (β-L-D4A), a potent and selective inhibitor of HBV replication, which is stronger than lamivudine (3TC)[3,4]. Because the nucleoside and nucleoside analog could serve as substrates for normal DNA polymerases, resulting in inhibition of cellular DNA synthesis, we decided to study the side effect of β-L-D4A on DNA polymerases[5].
It has been found that many kinds of DNA polymerase have a crucial role in DNA replication, among which, the high molecular weight class of eukaryotic DNA polymerase α (EC2.7.7.7), first identified in calf thymus extract and found to be ubiquitous among eukaryotes, constitutes the predominant polymerase species in actively dividing cells[6,7]. So ion exchange chromatography was used to separate DNA polymerase α from crude extract of human Hela cells, which could be used as a substrate to evaluate the safety of nucleoside-β-L-D4A.
β-L-D4 triphosphate was chemically synthesized in our institute with the help of Pharmaceutic College of Wuhan University and identified by infrared mass spectra, nuclear-magnetic resonance. 3TC 5' triphosphate was provided by Professor Cheng YC (School of Medicine, Yale University, New Haven, CT). Hela S3 cells were purchased from Institute of Immunity, Tongji Medical College, DE52 and P11 cellulose resin were from Whatman while HTP was from Bio-Rad. DNA-cellulose and activator calf thymus DNA were obtained from SIGMA. α-32P-dTTP was from Amersham. All other tissue culture reagents were from Gibco.
All operations were performed at 0°C-4°C. Unless otherwise noted, all buffers contained 1 mmol/L β-mercaptoethanol and 1 mmol/L EDTA.
Frozen cells (approximately 80 g) were suspended in 300 mL of 2 mmol/L MgCl2, 1 mmol/L p-toluenesulfonyl fluoride, 10 mL/L isopropyl alconol, and 5 mmol/L KPO4 (pH7.5), and broken in a Dounce homogenizer after 20 min at 0°C. Nuclei were spun down at 800 r/min for 10 min, and mitochondria were pelleted at 13 000 r/min for 20 min. The mitochondrial pellet was washed with 80 mL extraction buffer and re-sedimentated with the two supernatants combined (a). Fraction a was dialyzed for 8-12 h against 10 volumes twice with 0.2 mol/L sodium acetate (pH5.5) each time, after which a flocculent precipitate was removed by centrifugation at 13 000 r/min for 20 min (b). Fraction b was over-layered with 117 mL/L sucrose in 0.2 mol/L KPO4 (pH8.2) and spun for 90 min at 40 000 r/min in a Beckman rotor. The supernatant was saved (c).
Fraction c was diluted to a final concentration of 0.1 mol/L KPO4 at 1:1 with 1 mmol/L β-mercaptoethanol, 1 mmol/L EDTA and loaded onto a DEAE-cellulose column (5 cm × 20 cm ) equilibrated with the same buffer. The column was washed with 3 volumes of equilibration buffer, and the adsorbed activity was then eluted in a single step with 0.2 mol/L KPO4 (pH8.2). All the active fractions from the step were pooled (d).
Fraction d was diluted to a final concentration of 0.1 mol/L KPO4 with 1 mmol/L β-mercaptoethanol, 1 mmol/L EDTA and loaded onto a second DEAE-cellulose column (2.5 cm × 25 cm ). The column was washed with 3 volumes of equilibration buffer and then developed with a 5-6 column volume gradient from 0.1 mol/L to 0.2 mol/L KPO4 (pH8.2).
The peak fractions of protein obtained from DE52 were pooled and dialyzed against 300 g/L sucrose, 200 mL/L ethylene glycol, 0.1 mol/L KPO4 (pH7.2) (e). Fraction e was loaded onto a phosphocellulose column (2.5 cm × 20 cm). The column was developed with a 6-volume gradient from 0.1 mol/L to 0.3 mol/L KPO4 (pH7.2).
The peak fractions of DNA polymerase α from P11 were pooled and dialyzed against 300 g/L sucrose, 0.05 mol/L KPO4 (pH 6.8) (f). Fraction f was diluted at 1:1 with 1.0 mol/L KCl, 4 mL/L Triton X-100 and loaded onto a hydroxylapatite column (1 cm × 13 cm). The column was eluted with a 15-volume gradient from 0.025 mol/L to 0.2 mol/L KPO4 (pH7.2).
The peak fractions were pooled and concentrated by dialysis against 300 g/L sucrose, 0.1 mol/L KPO4 (pH7.5) (g). Fraction g was diluted to a final concentration of 0.02 mol/L KPO4 (pH7.5) at 1:4 with 1 mmol/L β-mercaptoethanol, 1 mmol/L EDTA, and loaded onto a DNA cellulose column (1 cm × 10 cm). The column was developed with a 20-volume gradient from 0.02 mol/L to 0.2 mol/L KPO4 (pH7.5).
Bradford method was used to measure the protein concentration, which was operated by directions of the reagent box.
The final volume of reaction mixture was 50 μL containing 2 μL DNA polymerase α, 50 μmol/L d CTP, 50 μmol/L d GTP, 50 μmol/L d ATP, 50 μmol/L α-32Pd TTP (100 cpm/pmol), 100 μg/mL activated calf thymus DNA, 50 mmol/L Tris-HCl (pH7.5), 0.5 mmol/L MnCl2, 100 mmol/L KCl and 2.5 mmol/L DTT. The mixture was Incubated at 37°C for 15 min, and spotted onto DE81 filter paper. The paper was washed three times with 5% Na2HPO4 (10 min each time), twice with water (5 min each time), dried and assayed for the acid-insoluble radioactivity.
SDS-PAGE was performed to detect the purity of DNA polymerase α.
Western blot was performed to confirm the protein bands.
Detailed kinetic parameters were determined using DNA polymerase α which was prepared, pooled and stored at -80°C until use in experiments. Potential inhibitors at various concentrations were added to 5 μL reaction mixture. Control samples (without inhibitor) included 5 μL solution in PBS, but no inhibitors. DNA polymerase α activity was measured.
To find the inhibition type, α-32P-dTTP(100 cpm/pmol) was performed at the concentration of 1, 3, 5, 7, 10 μmol/L while inhibitors were added at the concentration of 10 μmol/L of each compound. DNA polymerase α was determined. The data were analyzed with GraphPad Prism 4 Demo, which could finish Line weaver-Burk Plot and L-B linear secondary regression with the enzyme kinetics template.
To determine the concentration required to inhibit DNA polymerase α activity by IC50, initial experiments were designed with the gradient (1 μmol/L-400 μmol/L) of each compound. The concentration of α-32P-dTTP (100 cpm/pmol) was 3 μmol/L. All experiments were performed twice. Inhibition rate = (1 - count of drug class/blank control) × 100%. Half logarithm plot was used to process the data. DNA polymerase α activity in the control sample was set at 100%.
To find the inhibition constant (Ki), each compound was studied at five different inhibitor concentrations (10, 40, 100, 160, 200 μmol/L), and at various substrate concentrations (2, 5 μmol/L) in duplicate. Dixon plot was used to deal with the data.
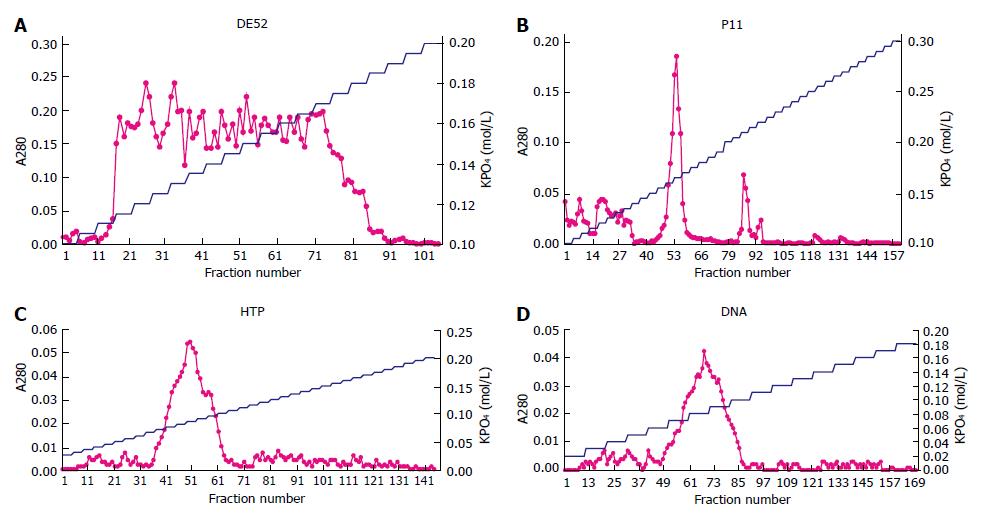
The first DE52 chromatography was to remove nucleic acid. The curve of A280 showed a broad peak including 74 tubes from the 16th tube (0.115 mol/L) to the 90th tube (0.185 mol/L) in the second DE52 chromatography (Figure 1A). The protein in 74 tubes was collected and the ingredient was confirmed by assay of DNA polymerase α activity. The total protein was 0.36 mg measured by Bradford method.
Protein determined by A280 was eluted sharply as a major peak at 0.165 mol/L, with a minor trailing shoulder occasionally present at 0.21 mol/L (Figure 1B). Fractions from 0.15 mol/L to 0.18 mol/L KPO4 were pooled because the minor peak was not the protein as expected. The total protein was 0.039 mg.
Fractions from the 37th to the 64th tube were collected because a single sharp peak appeared at 0.085 mol/L KPO4 (Figure 1C). The characterization was certified by its activity and the protein was 0.008 mg.
Protein was eluted in a single sharp peak at 0.085 mol/L KPO4 (Figure 1D). As the activity was measured, the region from 0.07 mol/L to 0.10 mol/L KPO4 was pooled and concentrated by dialysis against 300 g/L sucrose, 0.1 mol/L KPO4 (pH7.5).
When the specific activity became higher, the total protein, total activity and yield became lower in each stage, illustrating that DNA polymerase α was purified step by step while the other components were removed (Table 1).
| Step | Protein/mg | Activity/units | Specificactivity/(units/mg) | Purificationtimes | Yield(%) |
| Crude extract | 9.000 | 800 | 89 | ||
| First DEAE | 0.880 | 480 | 545 | 6 | 60 |
| Second DEAE | 0.360 | 260 | 722 | 8 | 33 |
| P11 | 0.039 | 240 | 6154 | 69 | 30 |
| HTP | 0.008 | 120 | 15000 | 169 | 15 |
| DNA | 0.002 | 32 | 31000 | 348 | 4 |
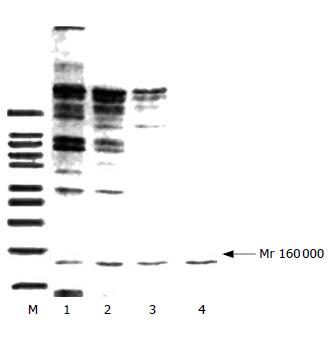
SDS-PAGE showed that the purification of protein was effective because the bands decreased with the step of purification and finally there was only one band (Figure 2). Western blot revealed that the band was DNA polymerase α (Figure 3).
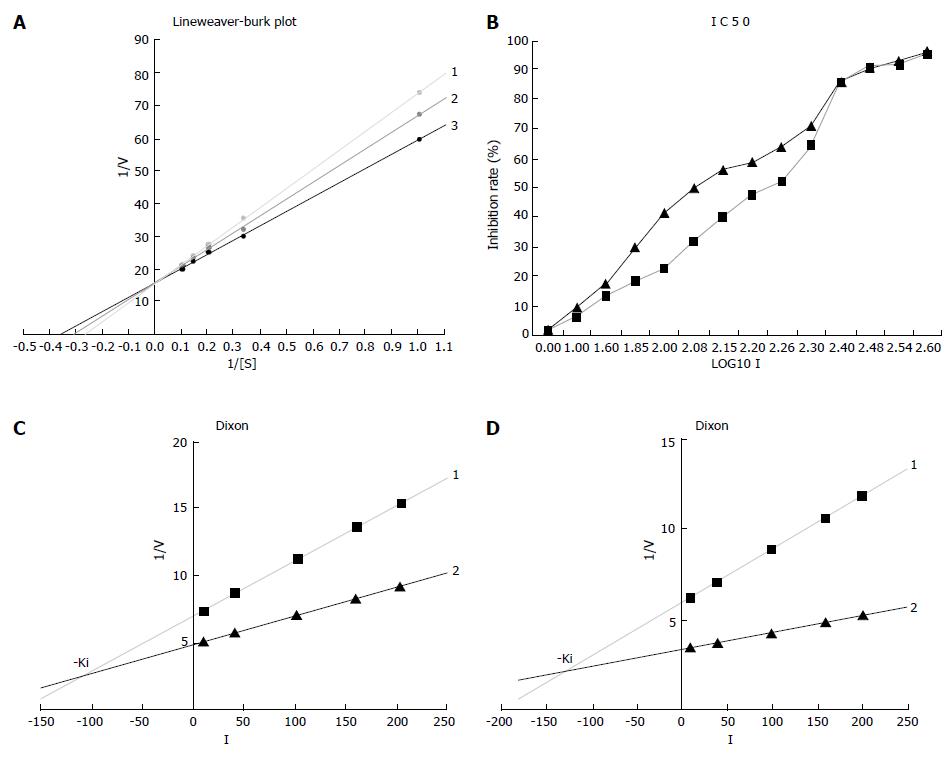
The inhibition mechanism was observed by visual inspection of graphical plots after data linearization (Line weaver-Burk plot) using the GraphPad Prism 4 Demo software. The L-B linear regression equations in Figure 4A showed 3TC5'-triphosphate, y = 15.922 + 57.978x (r2 = 0.9999), Km = 3.64 μmol/L; β-L-D4A, y = 15.879 + 51.163x (r2 = 0.9993), Km = 3.22 μmol/L; blank without inhibitor, y = 15.860 + 43.493x (r2 = 0.9995), Km = 2.74 μmol/L.
IC50 was calculated using the half logarithm plot, demonstrating that β-L-D4A (178.49 μmol/L) was 1.5 fold more potent than 3TC (122.41 μmol/L) (Figure 4B).
The Ki was also estimated using the GraphPad Prism 4 Demo software. The two linear regression equations in Figure 4C were 3TC, y = 0.0408x + 6.9594 (r2 = 0.9998) and y = 0.0212x + 4.8957 (r2 = 0.9991) while the equations in Figure 4D were β-L-D4A, y = 0.0297x + 6.0389 (r2 = 0.9965) and y = 0.0095x + 3.4904 (r2 = 0.9996). The Ki of 3TC was 105 μmol/L which was lower than β-L-D4A (126 μmol/L).
HBV infection, associated with the risk of developing liver cirrhosis and hepatocellular carcinoma, is a worldwide public health problem. Two billion people worldwide show evidence of having been infected with HBV, and more than 350 million of them are chronically infected. Persons with chronic hepatitis B are at a high risk of developing hepatic cirrhosis and primary hepatocellular carcinoma, leading to 500 000 to 1.2 million deaths annually worldwide. Antiviral chemotherapy remains the only choice of treatment for controlling HBV infection in these individuals, for whom available HBV vaccines provide no benefit. At present, approved therapeutics for chronic HBV infection are α-interferon or nucleoside and nucleoside analogs[8]. Drawbacks of treatment with α interferon include a low sustained response rate, undesirable side effects, the need for parenteral administration, and high cost[9,10]. Treatment with nucleosides such as 3TC is less costly and more convenient[11,12]. The fundamental concern is that while initial treatment of patients with 3TC results in a rapid decrease in HBV DNA blood levels, its efficacy is severely compromised in most patients by the development of antiviral resistance after prolonged therapy[13,14]. Although the use of nucleoside and nucleoside analogs as anti-hepadnaviral agents is similarly disappointing, prospects for their future use are bright, as several of recently developed analogs have been found to be potent and can be used as selective inhibitors of HBV replication. In our previous work, the novel nucleoside (β-L-D4A) was synthesized and its inhibitiory actions against HBV were studied and found to be much better than 3TC[15,16]. The inhibition of HBV replication by nucleoside analogs results from the recognition of nucleoside analog triphosphates (TPs) by the RNA-dependent DNA polymerase of HBV (HBV Pol). The triphosphate derivatives could serve as substrates for human DNA polymerases, inhibiting cellular DNA synthesis. For the systemic evaluation of the safety of the new drug, kinetic analysis of DNA polymerases must be performed.
Of the multiple DNA polymerases occurring in eukaryotes, only DNA polymerase α is able to initiate the synthesis of new strands[6], because it can initiate the replication of DNA by cooperating with RNA polymerase[17-19].
In the present study, DNA polymerase α was purified step by step because the total protein and activity decreased while the specific activity raised and the number of protein bands was cut down till only one band was left in SDS-PAGE (Mr 160 000). Western blot and the specific activity (31 000 units/mg) confirmed that the scheme was effective. The enzyme obtained lays a foundation for the next research.
Nucleoside analogs are chemically synthesized drugs that are able to mimic natural nucleosides[20,21]. They exert their antiviral effect, after anabolism to the triphosphate form, by acting as alternate substrates for the virally encoded reverse transcriptase[22,23]. Incorporation of the nucleoside analog monophosphate into the viral DNA, results in premature termination of viral DNA synthesis. Nucleoside analogs competitively inhibit DNA-dependent reverse transcriptase activity of the viral polymerase[24,25]. As DNA polymerase α has the similar substrate and mechanism to HBV viral polymerase, the drugs could serve as substrates for human DNA polymerase α, inhibiting cellular DNA synthesis[26,27]. Because DNA polymerase α exhibits neither exonuclease nor endonuclease activity, mistake cannot be repaired[28,29]. To determine the safety of nucleosides on cells in the present study, DNA polymerase α was purified and kinetic analysis was performed under the identical conditions of ionic strength, pH, divalent metal ion concentration, and DNA substrate.
The Km increased and Wmax unchanged (Figure 4A), confirming that both of β-L-D4A and 3TC are the competitive inhibitors of DNA polymerase α. In our study, the drugs inhibited DNA polymerase α activity by acting as competitive alternate substrates. 3TC was more effective than β-L-D4A as inhibitors of DNA polymerase α because IC50 of β-L-D4A was 1.5 fold more potent than 3TC and the Ki of 3TC was 105 μmol/L, much lower than β-L-D4A (126 μmol/L), demonstrating that β-L-D4A is significantly more safe than 3TC.
In conclusion, β-L-D4A is a safe drug for HBV infection because it is endowed with lower host toxicity in comparison to 3TC. Furthermore, combined therapy of β-L-D4A and lamivudine for HBV infection can be explored.
The authors thank Xiao-Dan Jiang, Institute of Immunity, Tongji Medical College.
Since hepatitis B virus (HBV) replication involves a virally encoded reverse transcriptase (RT), a number of L-configuration nucleoside analogs with an inhibitory effect on RT have emerged as potent antiviral agents against HBV infection. However, most anti-HBV nucleoside analogs tested to date have only transient and limited effects on a small number of HBV-infected individuals with moderate to severe side effects. In light of the fact, development of novel antiviral agents is an extremely important undertaking.
In recent years, a considerable interest has been focused on the use of 2', 3'-dideoxynucleosides (DDNs) in the treatment of chronic HBV infection. β-L-D4A, a novel L-nucleoside, can effectively block the production of HBV in 2.2.15 cells in vitro, but no information is available on the safety evaluation of this new compound.
In this research, kinetic analysis of DNA polymerase α cytotoxicity, was investigated to evaluate the safety of this new compound.
β-L-D4A is a safe drug for HBV infection because it is endowed with a low host toxicity.
β-L-D4A, [5-(6-amino-9H-purin-9-yl)-2,5-dihydrofuran-2-yl] methanol, is a novel L-nucleoside.
This is an interesting paper describing the effect of β-L-D4A (a novel nucleoside analog) on DNA polymerase α. The authors separated DNA polymerase α from crude extract of human Hela cells and studied its enzyme kinetics, showing that the enzyme is less inhibited than lamivudine, thus β-L-D4A may be an effective nucleoside for HBV infection with a lower host toxicity.
S- Editor Liu Y L- Editor Wang XL E- Editor Li HY
| 1. | Anderson RD, Chinnakotla S, Guo L, Perrillo RP, Klintmalm GB, Davis GL. Intramuscular hepatitis B immunoglobulin (HBIG) and nucleosides for prevention of recurrent hepatitis B following liver transplantation: comparison with other HBIG regimens. Clin Transplant. 2007;21:510-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Wu JM, Lin JS, Xie N, Liang KH. Inhibition of hepatitis B virus by a novel L-nucleoside, beta-L-D4A and related analogues. World J Gastroenterol. 2003;9:1840-1843. [PubMed] |
| 3. | Wu JM, Lin JS, Xie N, Jiang FC, Liang KH. [Effect and mechanism of beta-L-D4A (a novel nucleoside analog) against hepatitis B virus]. Zhonghua Ganzangbing Zazhi. 2003;11:268-270. [PubMed] |
| 4. | Wu JM, Lin JS, Xie N, Qiu GF, Hu XM. [Synthesis of a novel L-nucleoside, beta-L-D4A and its inhibition on the replication of hepatitis B virus in vitro]. Yaoxue Xuebao. 2005;40:825-829. [PubMed] |
| 5. | Oshige M, Takenouchi M, Kato Y, Kamisuki S, Takeuchi T, Kuramochi K, Shiina I, Suenaga Y, Kawakita Y, Kuroda K. Taxol derivatives are selective inhibitors of DNA polymerase alpha. Bioorg Med Chem. 2004;12:2597-2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Matsubara K, Saito A, Tanaka A, Nakajima N, Akagi R, Mori M, Mizushina Y. Epicatechin conjugated with fatty acid is a potent inhibitor of DNA polymerase and angiogenesis. Life Sci. 2007;80:1578-1585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Abdel-Aziz W, Hickey R, Edelman M, Malkas L. Effect of novel benzoylphenylurea derivatives on DNA polymerase alpha activity using the synthesome-based in vitro model system. Invest New Drugs. 2003;21:421-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 8. | Marcellin P, Lada O, Asselah T. Treatment of chronic hepatitis B with the combination of pegylated interferon with lamivudine. Hepatol Res. 2007;37:S55-S61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Kao JH. Appropriate use of interferon for treatment of chronic hepatitis B. Hepatol Res. 2007;37:S47-S54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 10. | Akyuz F, Kaymakoglu S, Demir K, Aksoy N, Karaca C, Danalioglu A, Onel D, Badur S, Besisik F, Cakaloglu Y. Lamivudine monotherapy and lamivudine plus interferon alpha combination therapy in HBeAg negative chronic hepatitis B not responding to previous interferon alpha monotherapy. Acta Gastroenterol Belg. 2007;70:20-24. [PubMed] |
| 11. | Lu HY, Zhuang LW, Yu YY, Ivan H, Si CW, Zeng Z, Li J, Hou DM, Chen XY, Han ZH. Intrahepatic HBV DNA as a predictor of antivirus treatment efficacy in HBeAg-positive chronic hepatitis B patients. World J Gastroenterol. 2007;13:2878-2882. [PubMed] |
| 12. | Hou J, Schilling R, Janssen HL, Hansen BE, Heijtink R, Sablon E, Williams R, Lau GK, Schalm SW, Naoumov NV. Genetic characteristics of hepatitis B virus genotypes as a factor for interferon-induced HBeAg clearance. J Med Virol. 2007;79:1055-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Palumbo E. Hepatitis B genotypes and response to antiviral therapy: a review. Am J Ther. 2007;14:306-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Sarin SK, Sood A, Kumar M, Arora A, Amrapurkar D, Sharma BC, Konar A, Chawla YK, Jain RK, Nanda V. Effect of lowering HBV DNA levels by initial antiviral therapy before adding immunomodulator on treatment of chronic hepatitis B. Am J Gastroenterol. 2007;102:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Wu Jm, Lin JS, Chen BT, Zheng XM, Zhao HB, Liang KH. [Establishment and identification of highly expressing and replicating hepatitis B virus genome transgenic mouse models]. Zhonghua Ganzangbing Zazhi. 2003;11:338-340. [PubMed] |
| 16. | Wu JM, Lin JS, Jiang FC, Zhang JY, Liang KH. [Inhibition of the replication of hepatitis B virus in vitro by a novel nucleoside analogue (beta-L-D4A)]. Zhonghua Ganzangbing Zazhi. 2003;11:446. [PubMed] |
| 17. | Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity. Biochemistry. 2007;46:448-460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Maeda N, Kokai Y, Ohtani S, Sahara H, Kuriyama I, Kamisuki S, Takahashi S, Sakaguchi K, Sugawara F, Yoshida H. Anti-tumor effects of dehydroaltenusin, a specific inhibitor of mammalian DNA polymerase alpha. Biochem Biophys Res Commun. 2007;352:390-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Christodoulou J, Craig HJ, Walker DC, Weaving LS, Pearson CE, McInnes RR. Deletion hotspot in the argininosuccinate lyase gene: association with topoisomerase II and DNA polymerase alpha sites. Hum Mutat. 2006;27:1065-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Caccamo L, Agnelli F, Reggiani P, Maggi U, Donato MF, Gatti S, Paone G, Melada E, Rossi G. Role of lamivudine in the posttransplant prophylaxis of chronic hepatitis B virus and hepatitis delta virus coinfection. Transplantation. 2007;83:1341-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Kawaoka T, Suzuki F, Akuta N, Suzuki Y, Arase Y, Sezaki H, Kawamura Y, Hosaka T, Kobayashi M, Ikeda K. Efficacy of lamivudine therapy in elderly patients with chronic hepatitis B infection. J Gastroenterol. 2007;42:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Uckun FM, Qazi S, Venkatachalam T. N'-[2-(2-thiophene) ethyl]-N'-[2-(5-bromopyridyl)]thiourea (HI-443), a rationally designed non-nucleoside reverse transcriptase inhibitor compound with potent anti-HIV activity. Arzneimittelforschung. 2007;57:278-285. [PubMed] |
| 23. | Parikh UM, Zelina S, Sluis-Cremer N, Mellors JW. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS. 2007;21:1405-1414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 24. | Fung J, Lai CL, Yuen JC, Wong DK, Tanaka Y, Mizokami M, Yuen MF. Adefovir dipivoxil monotherapy and combination therapy with lamivudine for the treatment of chronic hepatitis B in an Asian population. Antivir Ther. 2007;12:41-46. [PubMed] |
| 25. | Gish RG. Improving outcomes for patients with chronic hepatitis B. Curr Gastroenterol Rep. 2007;9:14-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 26. | Leemans W, Janssen HL, de Man R. Future prospectives for the management of chronic hepatitis B. World J Gastroenterol. 2007;13:2554-2567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | de Baar MP, de Rooij ER, Smolders KG, van Schijndel HB, Timmermans EC, Bethell R. Effects of apricitabine and other nucleoside reverse transcriptase inhibitors on replication of mitochondrial DNA in HepG2 cells. Antiviral Res. 2007;76:68-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Qi X, Xiong S, Yang H, Miller M, Delaney WE 4th. In vitro susceptibility of adefovir-associated hepatitis B virus polymerase mutations to other antiviral agents. Antivir Ther. 2007;12:355-362. [PubMed] |
| 29. | D'Ugo E, Kondili LA, Canitano A, Catone S, Giuseppetti R, Gallinella B, Palmieri G, Orobello S, Argentini C, Glück R. Rapid emergence of a viral resistant mutant in WHV chronically infected woodchucks treated with lamivudine and a pre-S/S CHO-derived hepatitis B virus vaccine. Vaccine. 2007;25:4895-4902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |