Copyright
©The Author(s) 2003.
World J Gastroenterol. May 15, 2003; 9(5): 1003-1007
Published online May 15, 2003. doi: 10.3748/wjg.v9.i5.1003
Published online May 15, 2003. doi: 10.3748/wjg.v9.i5.1003
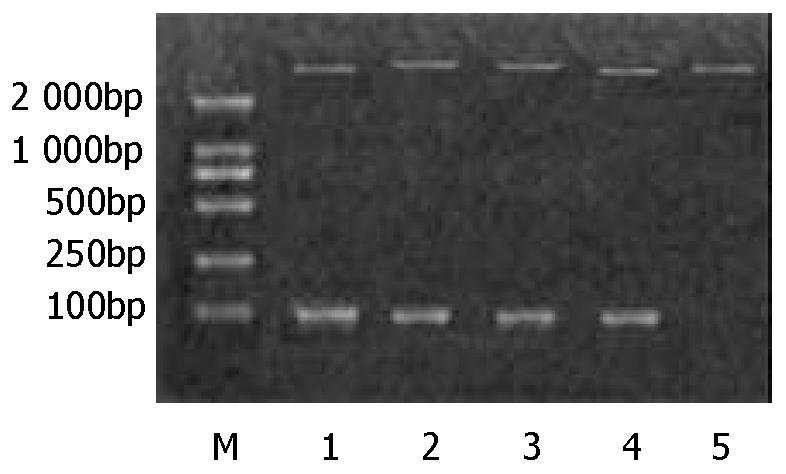
Figure 1 Digestion of the recombinant plasmids with Hind III and Bgl II.
M is DNA marker (DL 2000). Lane 1-lane 4 show four recombinant plasmid digestion results, SH1b, BJ1b, SD1b and SD2a respectively. Lane 5 is the control (pQE40).
Figure 2 A: The nucleotide sequences of the four HVR1 fragments in recombinant pQE40-HVR1 palsmids.
Dashes represent nucleotides identical to those of HVR1-SH1b. B: Deduced amino acid sequence of the HVR1 frag-ments in pQE40-HVR1. Sequences are shown with a single-letter amino acid code where residue is different from the con-sensus sequence of genotype 1b defined by Hattori et al[10], and with a dash where residue is identical.
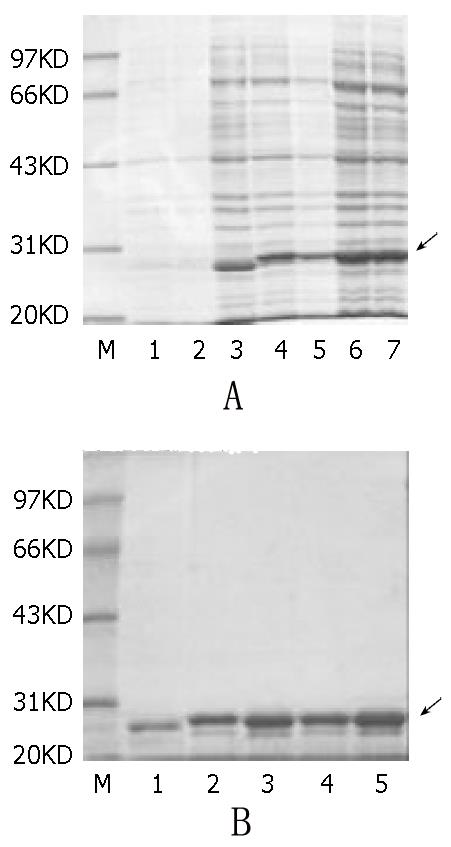
Figure 3 A: SDS-PAGE of the fusion proteins expressed in E.
coli strain TG1. M: protein molecular marker. Lane 1 is vector pQE40 before induction, Lane 2 is one of the recombinant plas-mids before induction. Lane 3-7, are recombinant plasmids pQE40-SH1b, BJ1b, SD1b, and SD2a after 4 hours of induction with 1 mmol/l IPTG respectively. B: Results of the purified proteins. M is protein marker. Lane 1 is plain DHFR. Lane 2-5 are the fusion proteins SH1b, BJ1b, SD1b and SD2a serially.
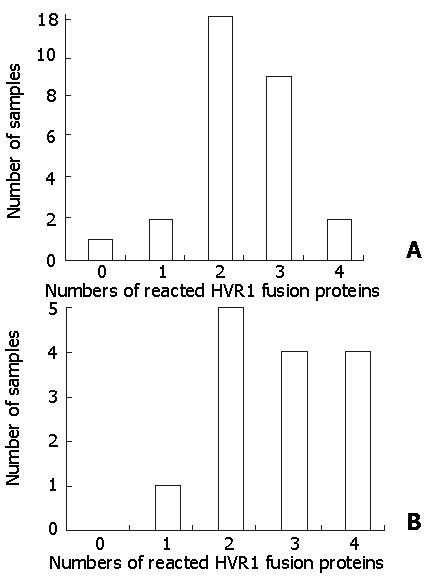
Figure 4 The serum of hepatitis C patients reacted with different numbers of HVR1 fusion proteins.
A shows that the sera of IFN responders reacted with 0-4 kinds of HVR1sequence; B shows that the sera of IFN non-responders reacted with 1-4 kinds of HVR1sequnce.
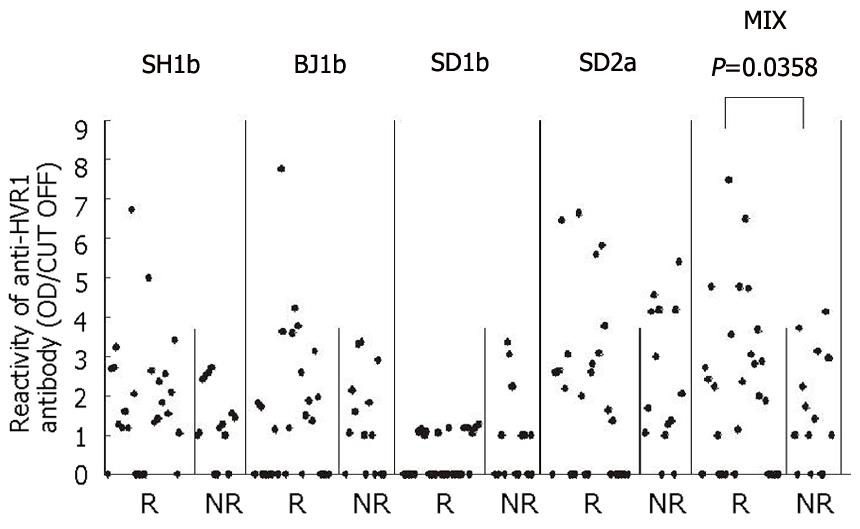
Figure 5 The OD values of 40 HCV-infected patients’ sera reacted with the four HVR1 fusion proteins respectively or totally, which were compared between R (IFN-responders) and NR (IFN-non responders).
For the mixed fusion proteins, the values (OD/CUT OFF) of the responders’ sera were significantly higher than those of non-responders (P = 0.0358).
- Citation: Zhang XX, Zhang SY, Liu J, Lu ZM, Wang Y. Expression of hepatitis C virus hypervariable region 1 and its clinical significance. World J Gastroenterol 2003; 9(5): 1003-1007
- URL: https://www.wjgnet.com/1007-9327/full/v9/i5/1003.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i5.1003