Copyright
©The Author(s) 2023.
World J Gastroenterol. Feb 21, 2023; 29(7): 1202-1218
Published online Feb 21, 2023. doi: 10.3748/wjg.v29.i7.1202
Published online Feb 21, 2023. doi: 10.3748/wjg.v29.i7.1202
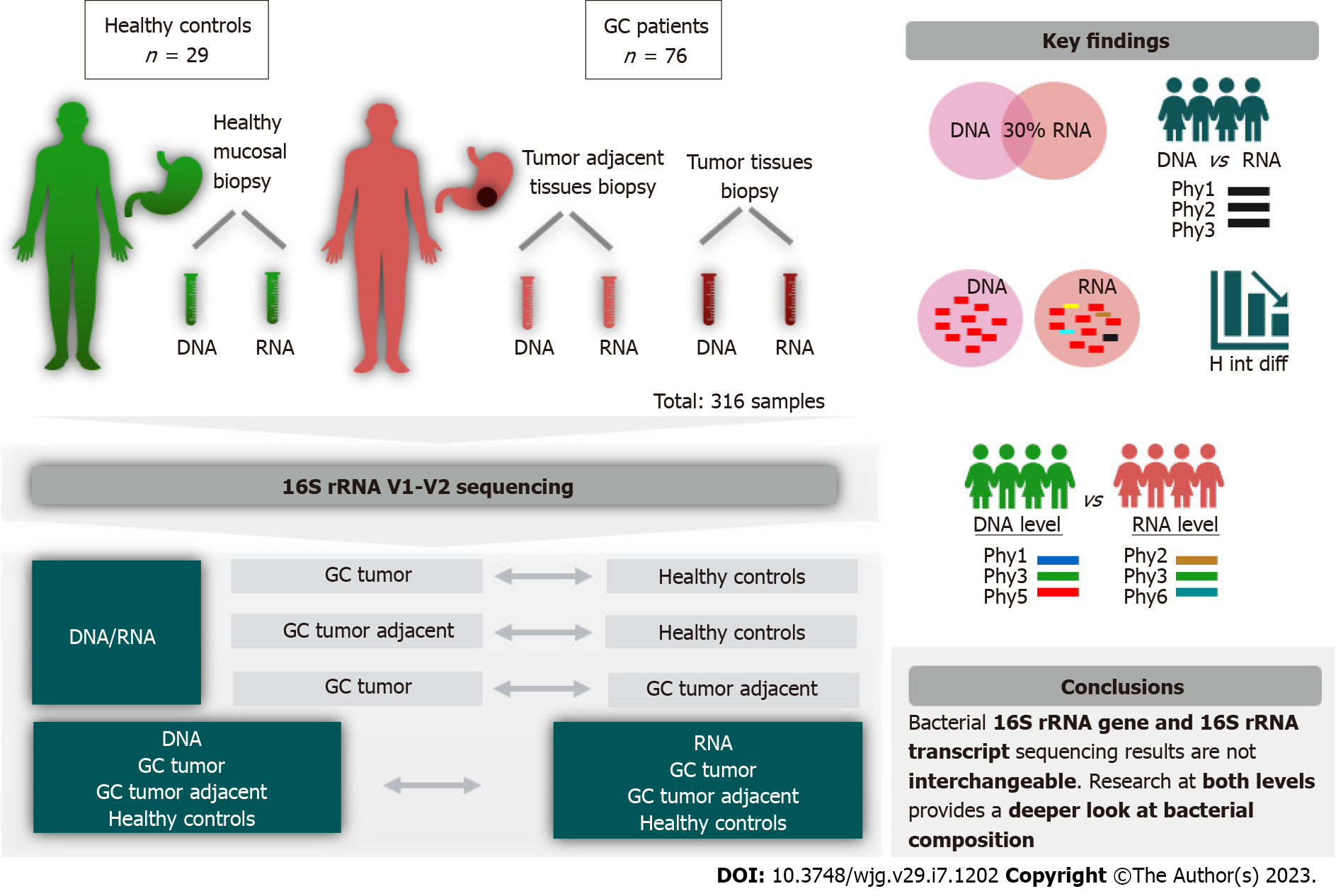
Figure 1 Study design illustration.
GC: Gastric cancer.
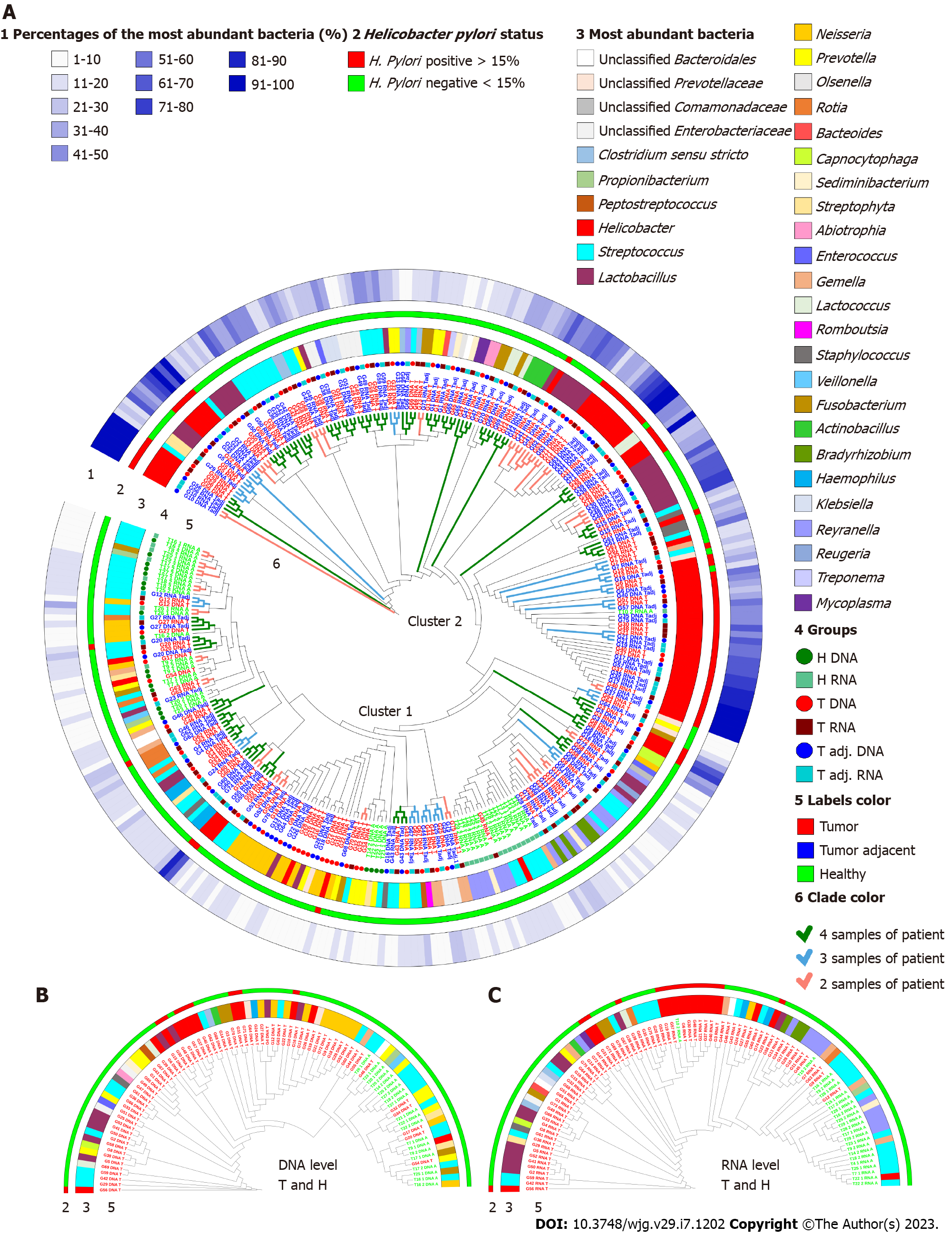
Figure 2 Group-average agglomerative hierarchical clustering at the phylotype level based on global bacterial profiles.
A: Clustering of 139 gastric cancer (GC) tumor, 134 tumors adjacent and 43 control group samples according to individual and general bacterial profiles; B and C: Clustering of GC tumor tissues and the control group at the DNA and RNA levels. The first circle (1) depicts the part of the most abundant bacteria; the second circle (2) represents the status of Helicobacter pylori; the third circle (3) shows the most abundant bacteria at the genus level; fourth and fifth circles (4 and 5) display the samples belonging to a certain study group; clade colors (6) represent the abundance of each patient sample clustered together. H. pylori: Helicobacter pylori.
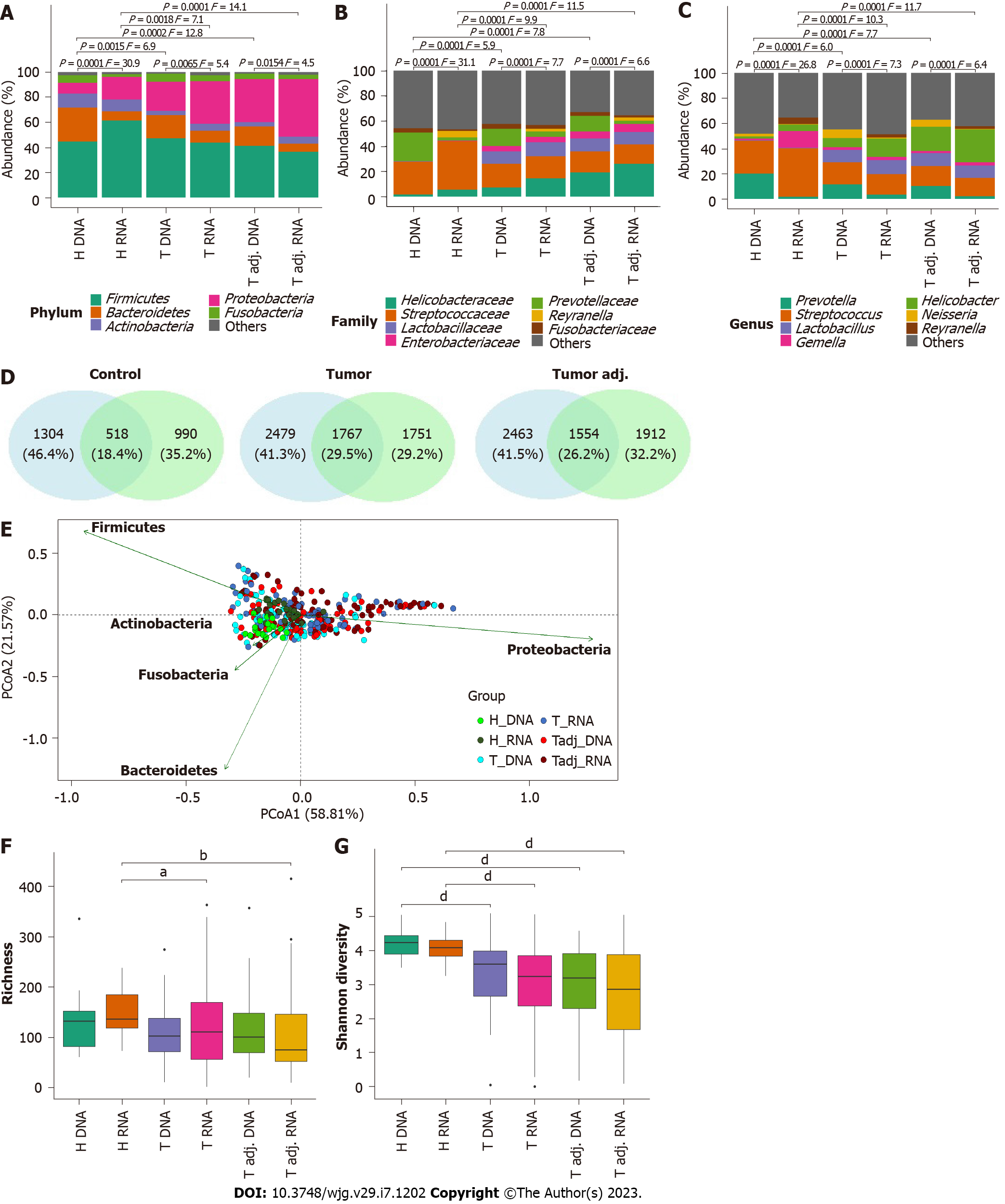
Figure 3 Shift and diversity of stomach microbiota of healthy controls, gastric cancer patient’s tumor and tumor adjacent tissues depending on microbial source (DNA and RNA).
A: Relative mean abundance of the top 5 microbiota at the phylum; B: Top 7 microbiota at the family; C: Top 7 microbiota at the genus level. Formal comparisons between groups were evaluated by analysis of similarity (PERMANOVA); D: Venn diagrams show the number of common and group-specific phylotypes comparing each study group at the DNA and RNA levels; E: Principal coordinate analysis of bacterial beta diversity of all study groups at the phylum level; F and G: Comparison of bacterial alpha diversity: Bacterial richness and bacterial diversity. aq < 0.05, bq < 0.01, dq < 0.0001.
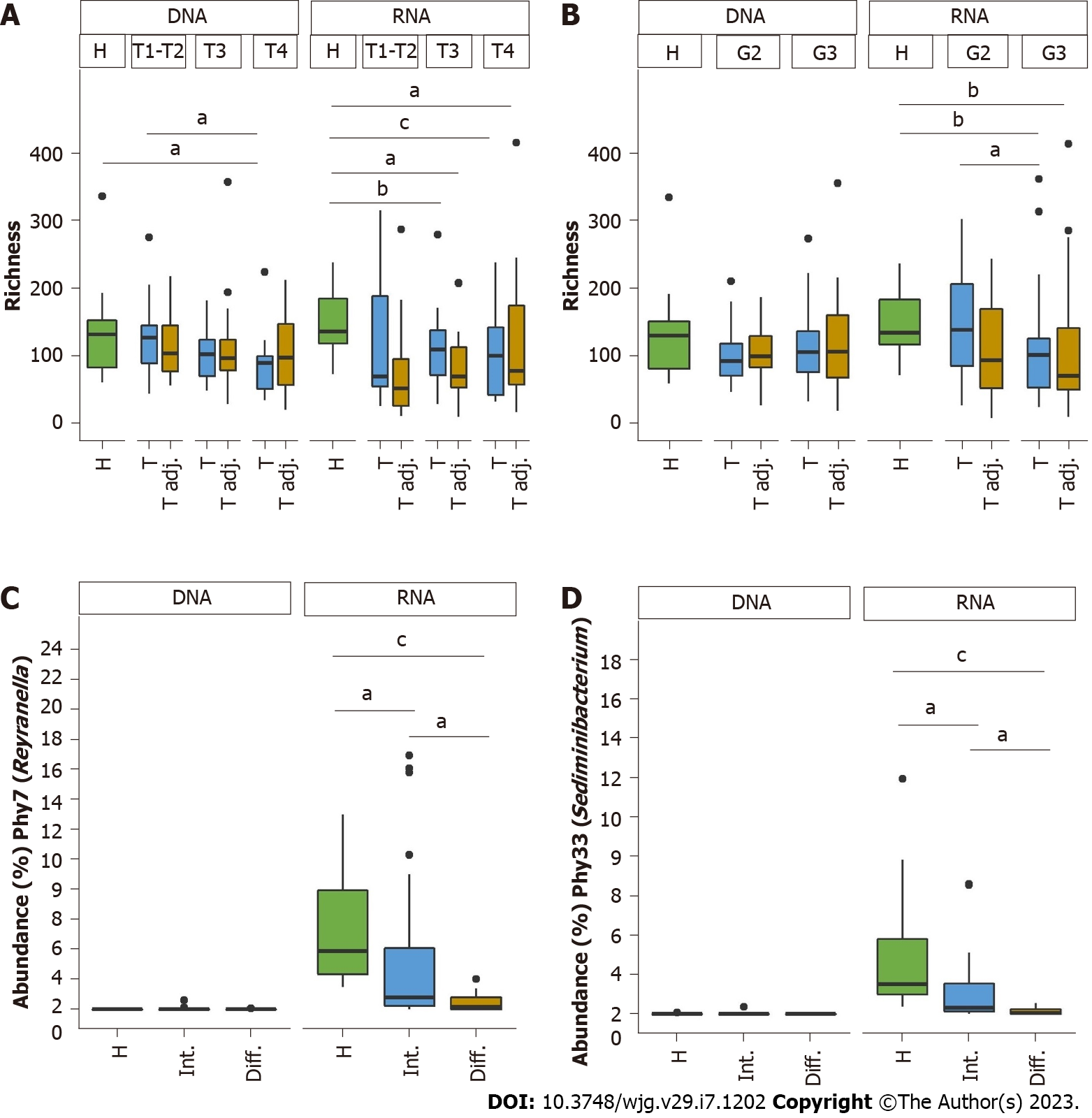
Figure 4 Comparison of bacterial abundance and richness in relation to clinical data at the DNA and RNA levels.
A: Bacterial richness according to tumor size; B: Bacterial richness according to grading; C and D: Comparison of the relative mean abundance of Phy7 (Reyranella) and Phy33 (Sediminibacterium). Statistically significant differences between study groups are indicated, aq < 0.05, bq < 0.01, cq < 0.001.
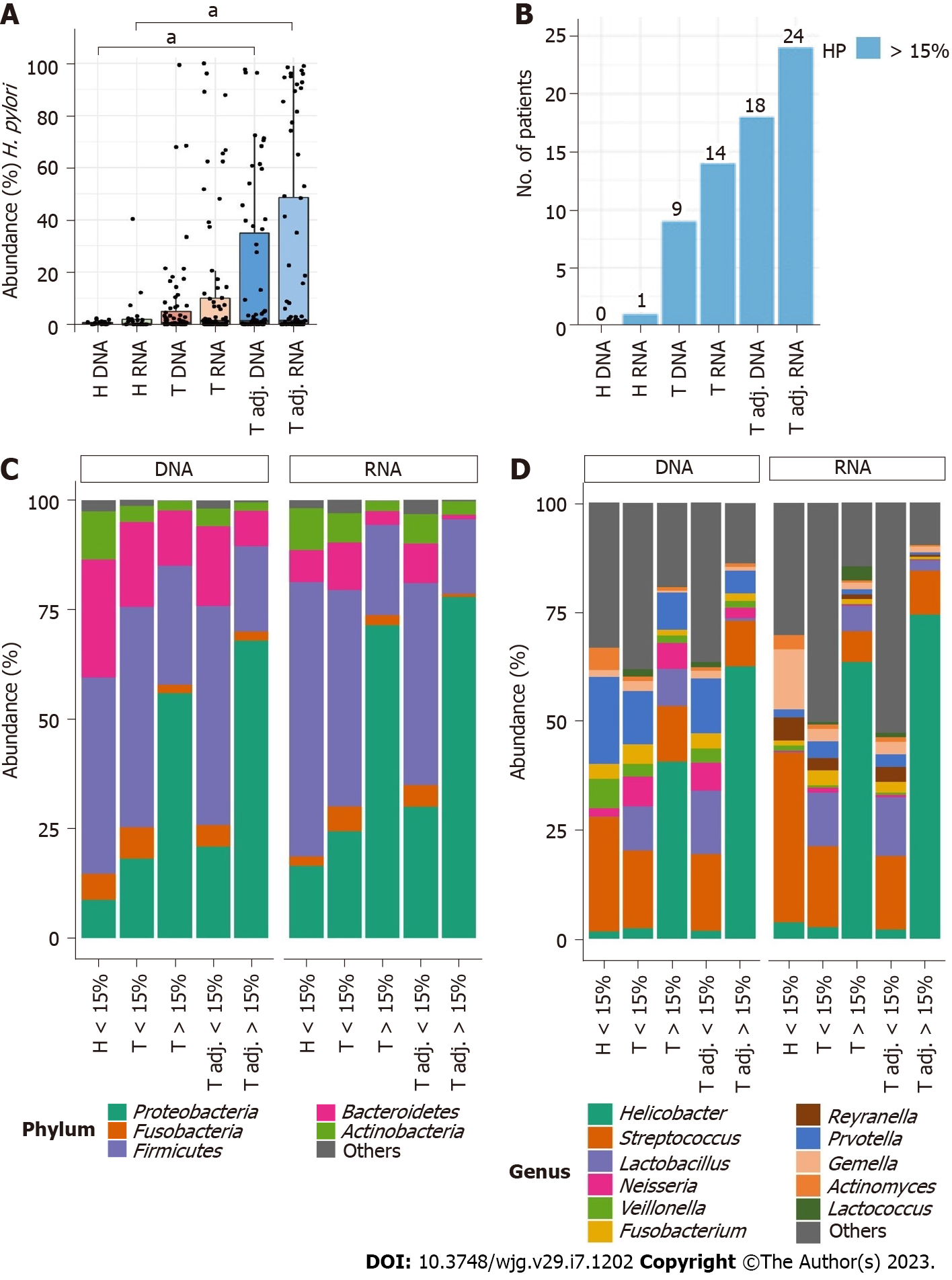
Figure 5 Comparison of gastric microbiota alterations at the DNA and RNA levels depending on high Helicobacter pylori abundance (> 15%).
A and B: Relative mean abundance of Helicobacter pylori (H. pylori) in all study groups and the number of patients with high H. pylori abundance; C and D: Comparison of the relative abundance of the top 5 microbiota at the phylum and top 11 microbiota at the genus levels. aq < 0.05.
- Citation: Nikitina D, Lehr K, Vilchez-Vargas R, Jonaitis LV, Urba M, Kupcinskas J, Skieceviciene J, Link A. Comparison of genomic and transcriptional microbiome analysis in gastric cancer patients and healthy individuals. World J Gastroenterol 2023; 29(7): 1202-1218
- URL: https://www.wjgnet.com/1007-9327/full/v29/i7/1202.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i7.1202