Copyright
©The Author(s) 2023.
World J Gastroenterol. Jun 14, 2023; 29(22): 3469-3481
Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3469
Published online Jun 14, 2023. doi: 10.3748/wjg.v29.i22.3469
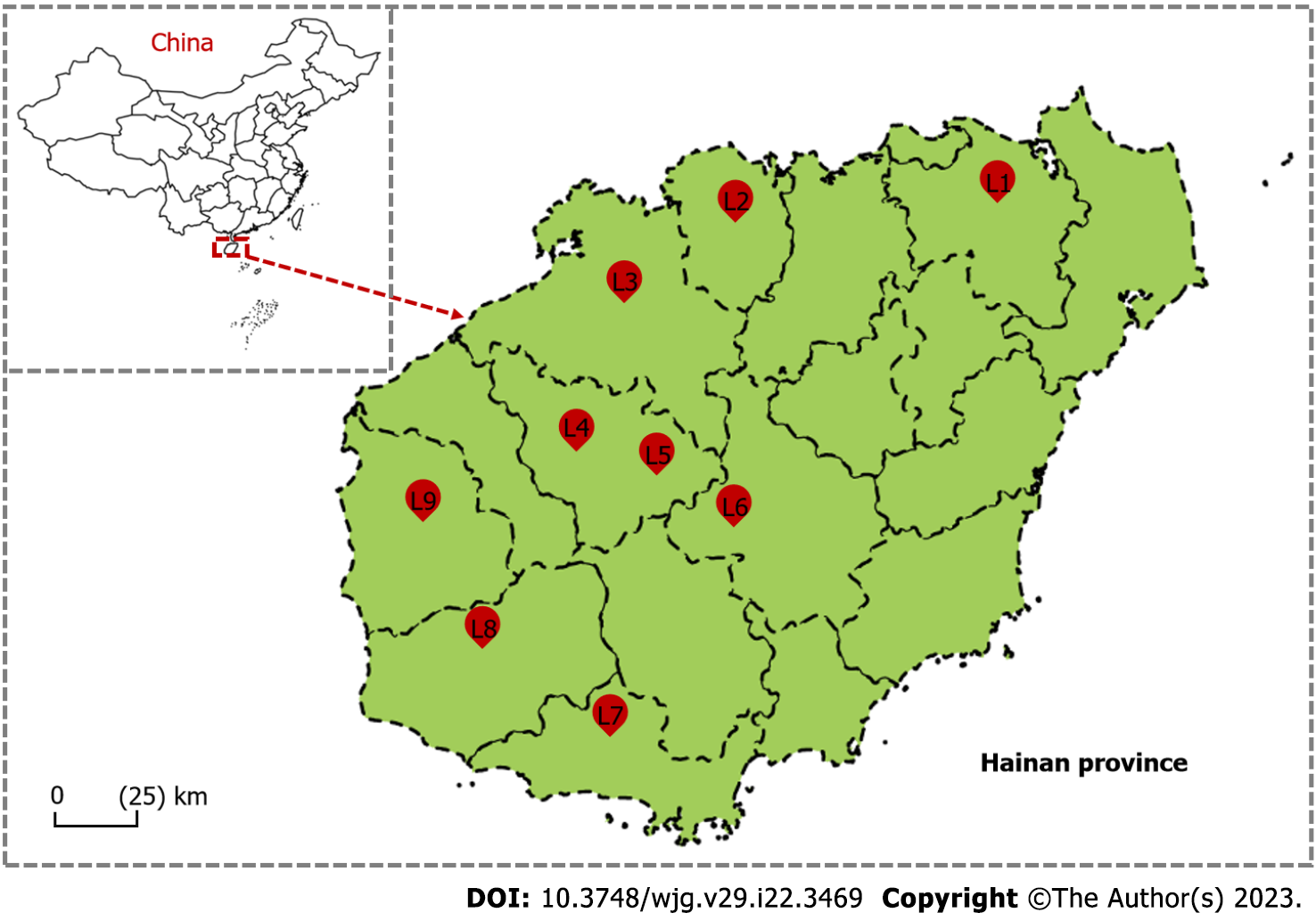
Figure 1 Sketch map of Hainan province.
The map shows the locations of the nine collection study sites. L1 = Yinggeling, mountain; L2 = Huangjingjiaoling, mountain; L3 = Danzhou, residential areas; L4 = Dongfang, residential areas; L5 = Haikou, residential areas; L6 = Jianfengling, mountain; L7 = Baisha, residential areas; L8 = Sanya, residential areas; L9 = Lingao, farmland.
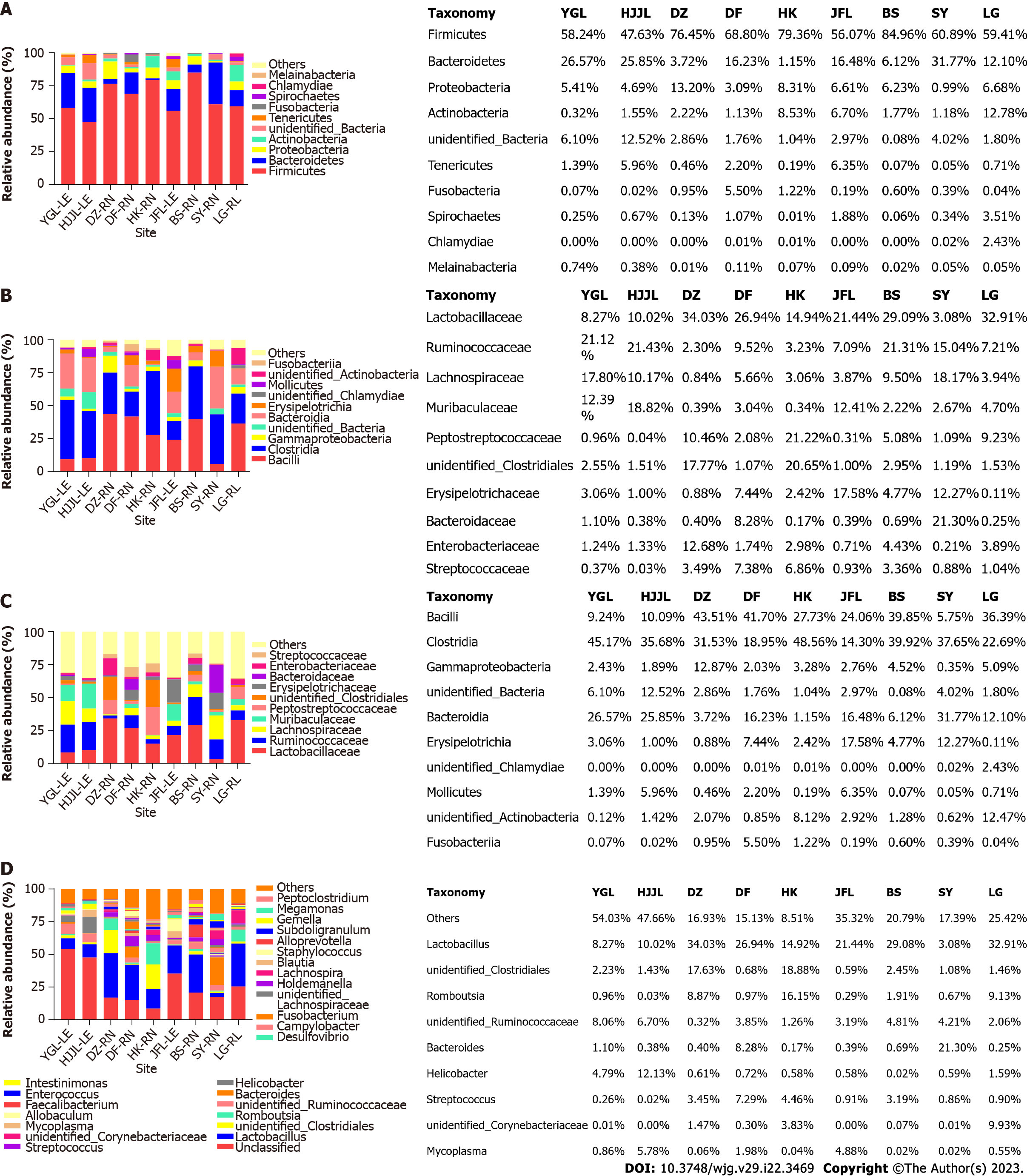
Figure 2 Mean relative abundances of bacterial phylum, class, family, and genus associated with three rat species at different sites.
A: Phylum; B: Class; C: Family; D: Genus. Families with an abundance of less than 2.50% and Phyla with an abundance of less than 0.15% were combined to form the category "Others." YGL: Yinggeling; HJJL: Huangjingjiaoling; DZ: Danzhou; DF: Dongfang; HK: Haikou; JFL: Jianfengling; BS: Baisha; SY: Sanya; LG: Lingao; LE: Leopoldamys edwardsi; RN: Rattus norvegicus; RL: Rattus losea.
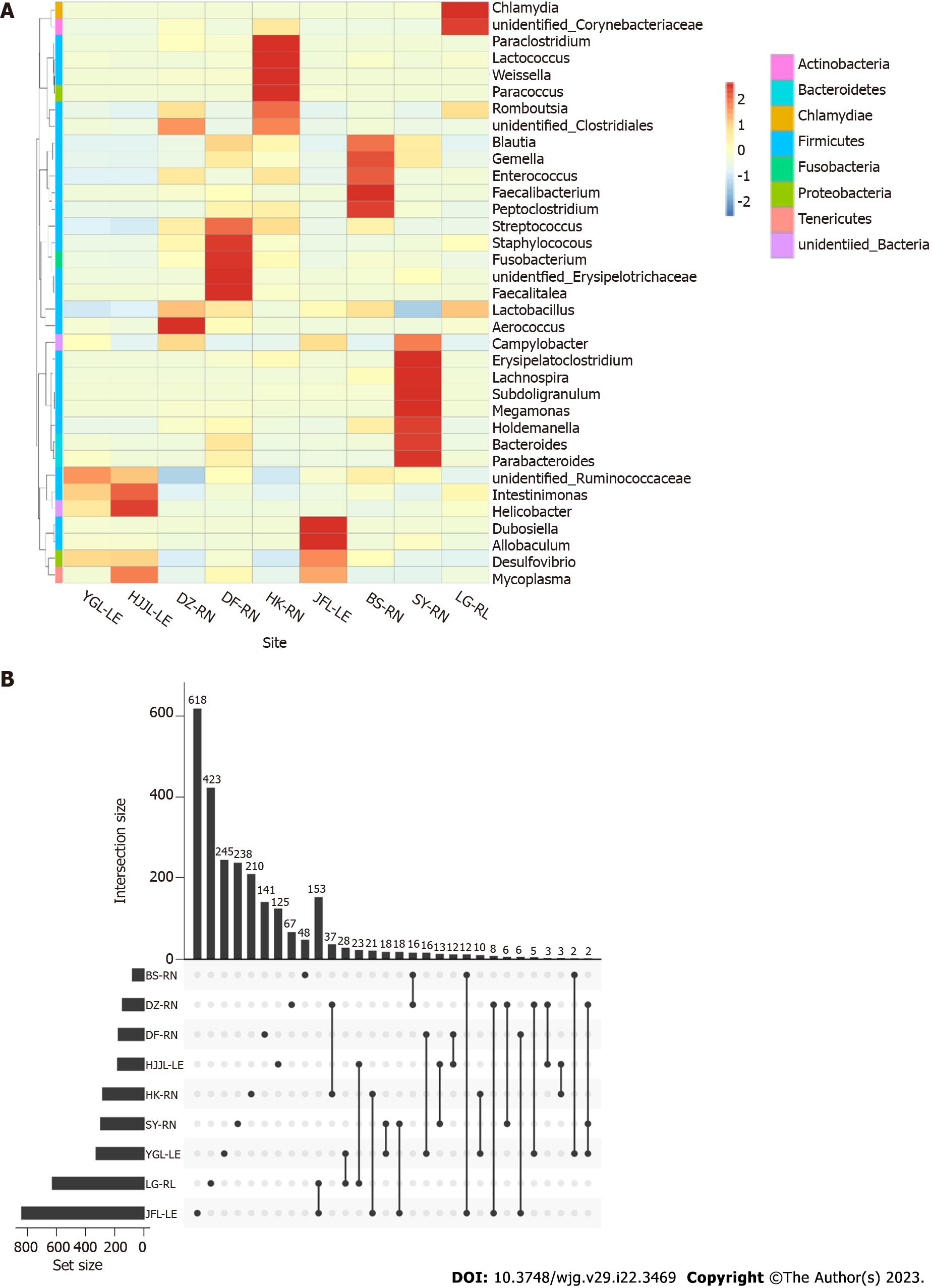
Figure 3 Heatmap and upset plot featuring the rat gut bacterial community.
A: The heatmap in log scale depicting the gut bacterial community of rat feces obtained with open reference operational taxonomic unit (OTU) picking methods. Blue colors represent high abundance, and red colors represent low abundance; white indicates absence; B: Visualizing the relationships between OTUs in all rat species across all sites using an upset plot. The Sangerbox tools, a free online platform for data analysis, were used to create the plot (http://www.sangerbox.com/tool). YGL: Yinggeling; HJJL: Huangjingjiaoling; DZ: Danzhou; DF: Dongfang; HK: Haikou; JFL: Jianfengling; BS: Baisha; SY: Sanya; LG: Lingao; LE: Leopoldamys edwardsi; RN: Rattus norvegicus; RL: Rattus losea.
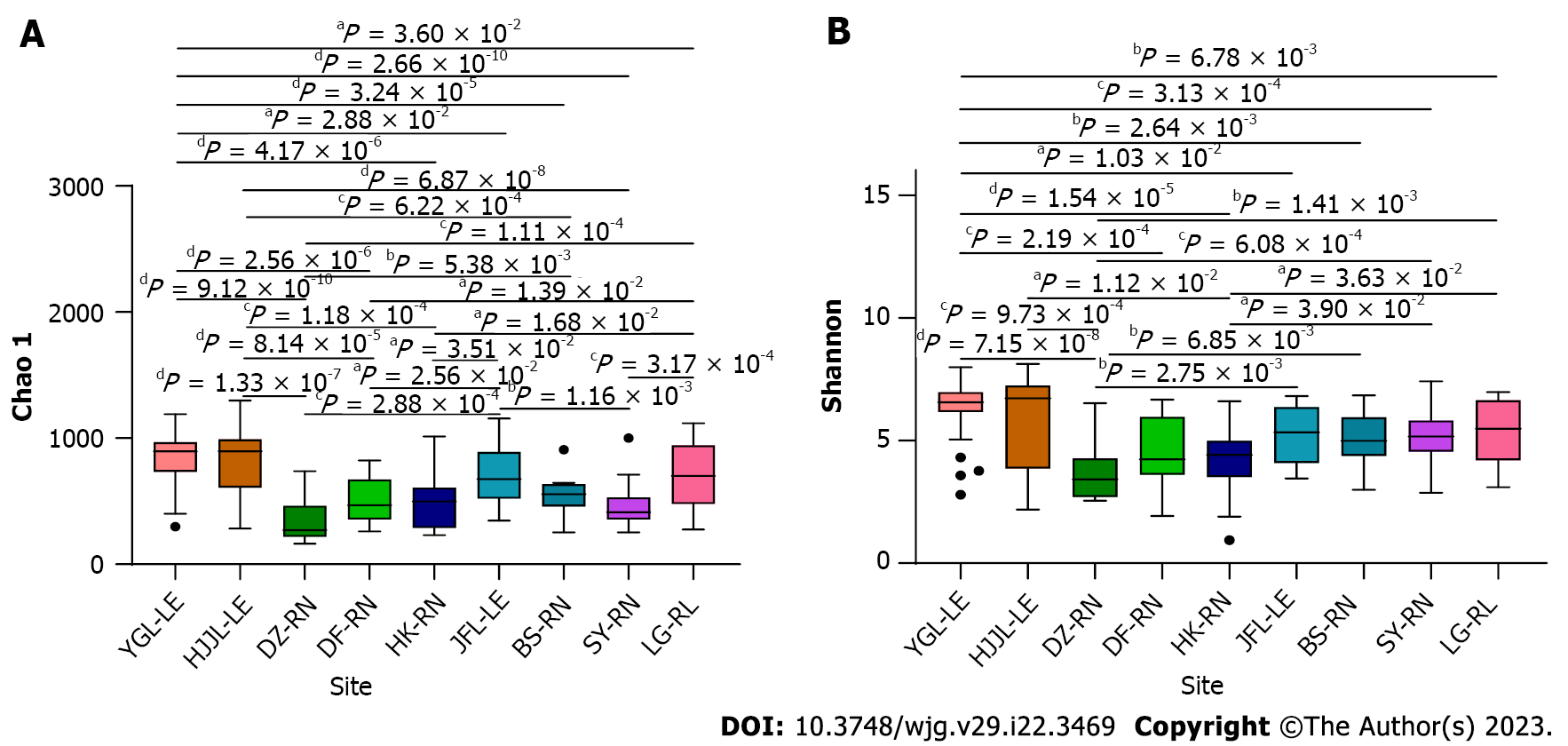
Figure 4 Boxplot representation of observed species.
Boxplots show the distribution of bacteria between rat samples categorized under different locations and rat species. A: Boxplot representation of chao1; B: Boxplot representation of Shannon diversity. Values from all statistical tests are available in additional Table 2. YGL: Yinggeling; HJJL: Huangjingjiaoling; DZ: Danzhou; DF: Dongfang; HK: Haikou; JFL: Jianfengling; BS: Baisha; SY: Sanya; LG: Lingao; LE: Leopoldamys edwardsi; RN: Rattus norvegicus; RL: Rattus losea. aP < 0.05; bP < 0.01; cP < 0.001; dP < 0.0001.
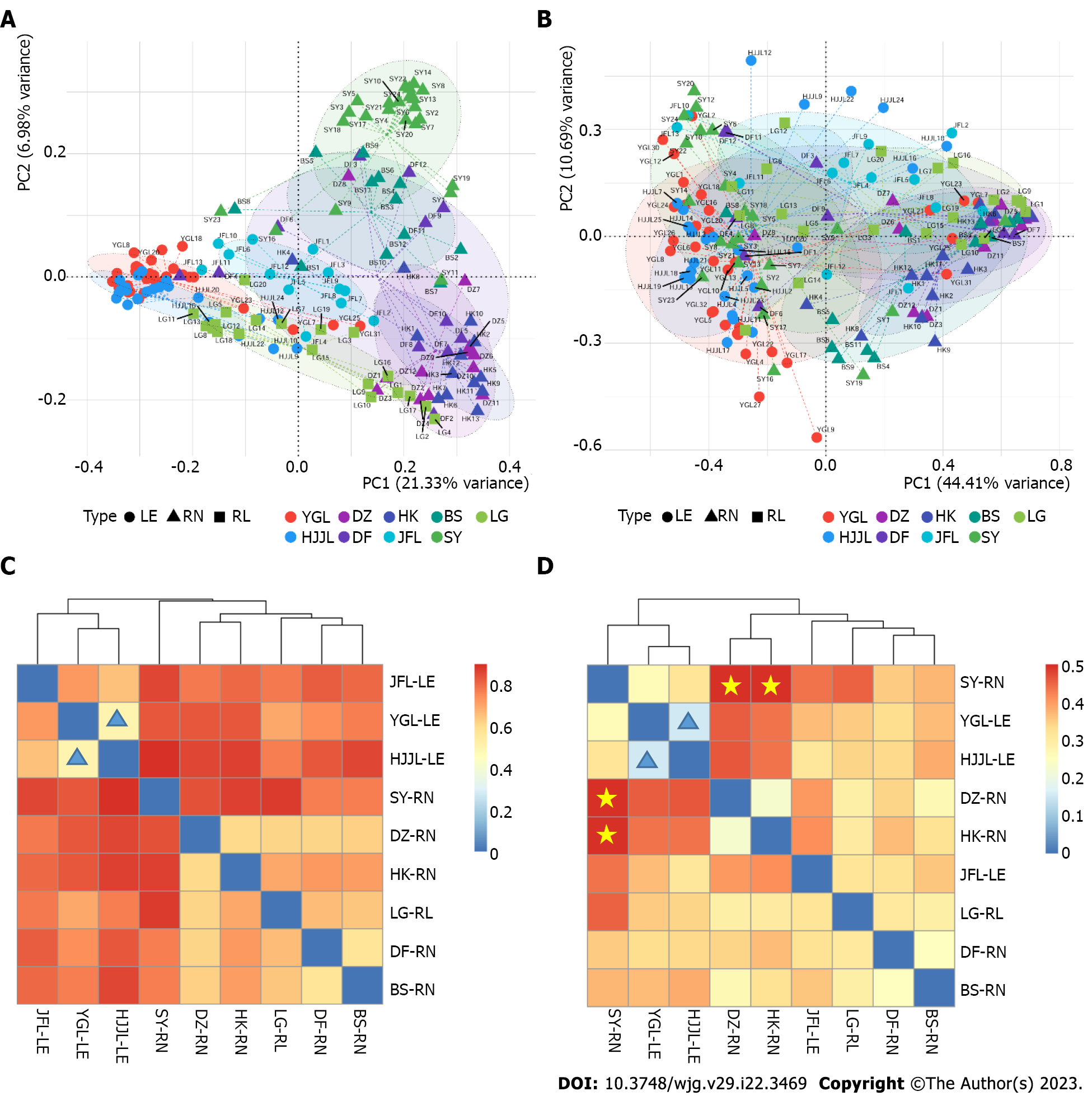
Figure 5 Principal coordinate analysis plot based on unweighted Unifrac distance and weighted Unifrac distance matrix depicting differences in the composition of the gut microbiota from different rat species and locations.
A: unweighted Unifrac distance; B: Weighted Unifrac distance; C and D: Colors represent community profiles of location. Matrix heatmap of the Bray-Curtis distances (C) and the weighted unifrac beta diversity (D) between the microbial communities of rats from nine different areas. YGL: Yinggeling; HJJL: Huangjingjiaoling; DZ: Danzhou; DF: Dongfang; HK: Haikou; JFL: Jianfengling; BS: Baisha; SY: Sanya; LG: Lingao; LE: Leopoldamys edwardsi; RN: Rattus norvegicus; RL: Rattus losea.
- Citation: Niu LN, Zhang GN, Xuan DD, Lin C, Lu Z, Cao PP, Chen SW, Zhang Y, Cui XJ, Hu SK. Comparative analysis of the gut microbiota of wild adult rats from nine district areas in Hainan, China. World J Gastroenterol 2023; 29(22): 3469-3481
- URL: https://www.wjgnet.com/1007-9327/full/v29/i22/3469.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i22.3469