Copyright
©The Author(s) 2023.
World J Gastroenterol. Jan 7, 2023; 29(1): 96-109
Published online Jan 7, 2023. doi: 10.3748/wjg.v29.i1.96
Published online Jan 7, 2023. doi: 10.3748/wjg.v29.i1.96
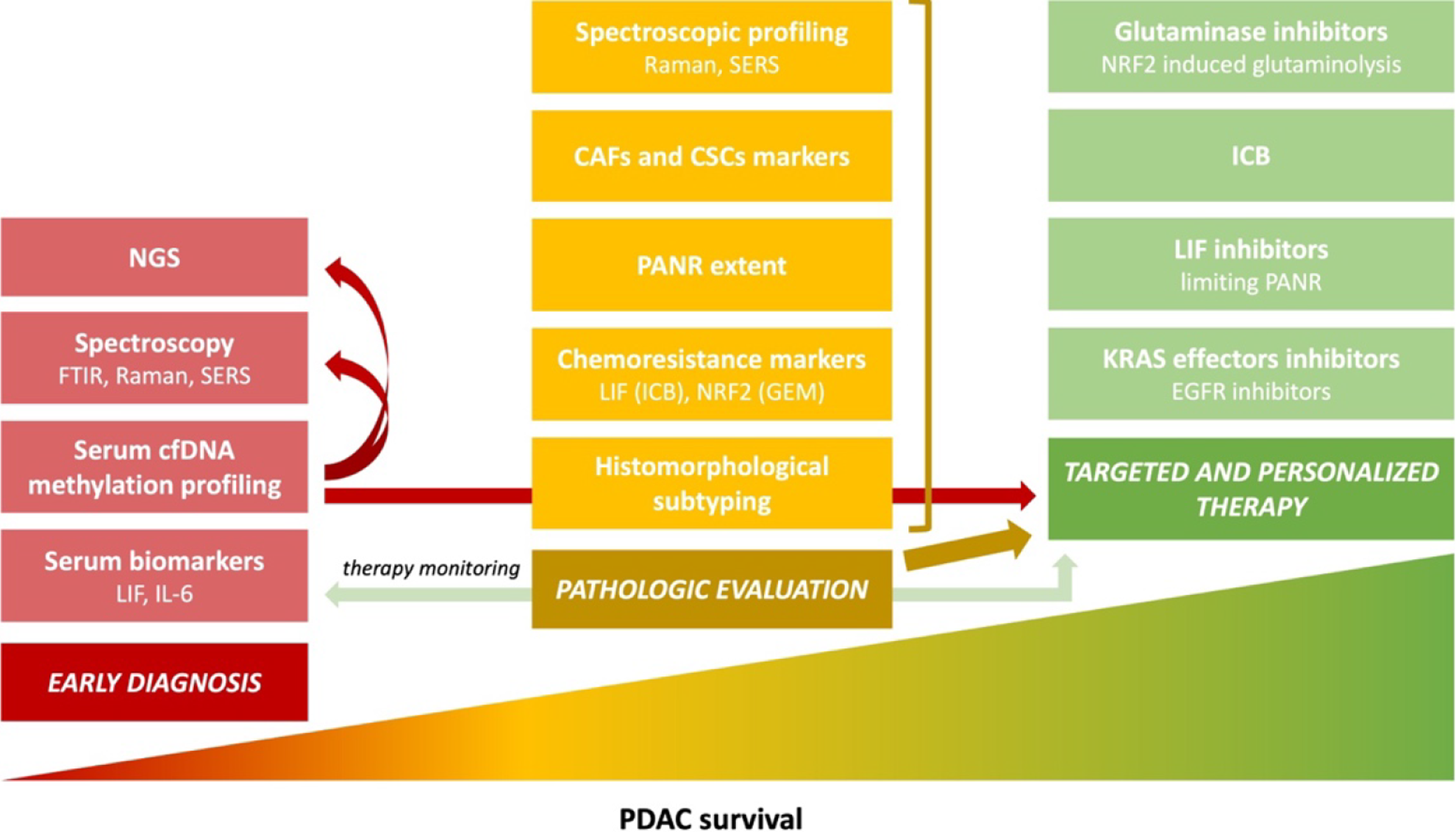
Figure 1 Main trends in pancreatic cancer pathology and research aim to improve survival[3].
Poor pancreatic ductal adenocarcinoma (PDAC) prognosis for a patient is multifactorial, and the lack of sensitive and specific early diagnostic methods is one of the reasons. Another is the resistance to available therapeutic options, which is caused, among others, by the tumor's molecular and morphological heterogeneity. Detailed pathological reporting is crucial for targeted and personalized therapy. The development of new diagnostic methods, combined with a proper pathologic evaluation and spectroscopic profiling, lead to effective treatment and supposedly will increase PDAC patients’ survival rates; Adapted with permission from[3]. Citation: Szymoński K, Milian-Ciesielska K, Lipiec E, Adamek D. Current Pathology Model of Pancreatic Cancer. Cancers (Basel) 2022; 14: 2321. Copyright© The Authors 2020. Published by MDPI. The image may be redistributed without special permissions–source: https://www.mdpi.com/openaccess. PDAC: Pancreatic ductal adenocarcinoma; LIF: Leukemia inhibitory factor; IL-6: Interleukin-6; cfDNA: Cell-free DNA; FTIR: Fourier transform infrared spectroscopy; Raman: Raman spectroscopy; SERS: Surface-enhanced Raman spectroscopy; NGS: Next-generation sequencing; ICB: Immune checkpoint blockers; NRF2: Nuclear factor-erythroid factor 2-related factor 2; GEM: Gemcitabine; PANR: PDAC-associated neural remodeling; CAFs: Cancer-associated fibroblasts; CSCs: Cancer stem cells; EGFR: Epithelial growth factor receptor.
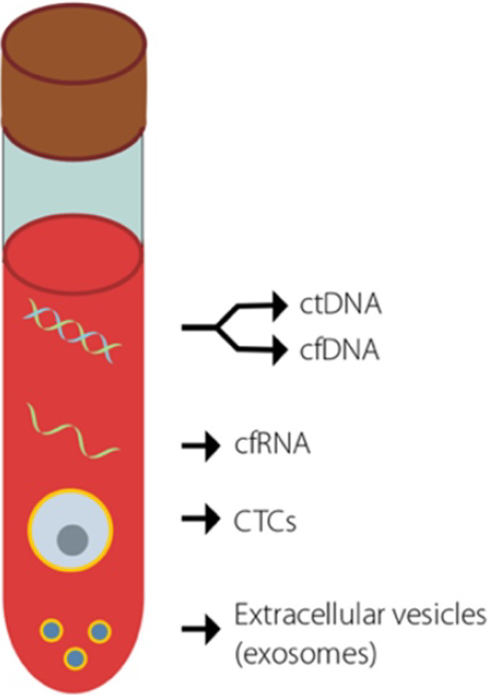
Figure 2 Potential cancer biomarkers that are detectable using liquid biopsies[16].
Key biomarkers that are currently used in an attempt to detect early-stage cancer. Adapted with permission from[16]. Citation: Jaworski JJ, Morgan RD, Sivakumar S. Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer. Cancers (Basel) 2020; 12: 3704. Copyright© The Authors 2020. Published by MDPI. The image may be redistributed without special permissions–source: https://www.mdpi.com/openaccess. ctDNA: Circulating tumor DNA; cfDNA: Cell-free DNA; cfRNA: Cell-free RNA; CTCs: Circulating tumor cells.
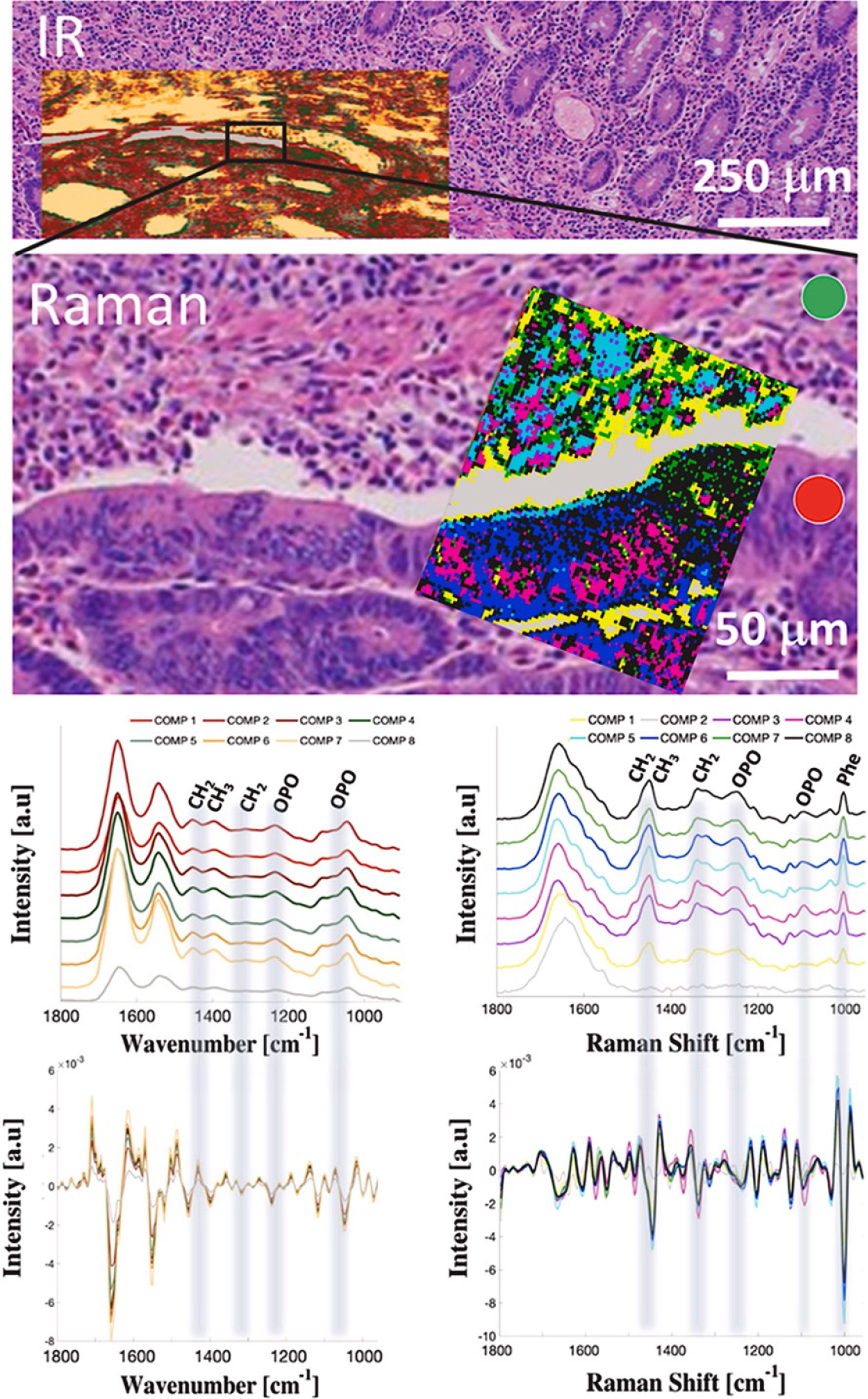
Figure 3 Spectroscopic mapping of ampullary adenocarcinoma[34].
A hematoxylin-eosin slide image of ampullary cancer tissue with superimposed Fourier transform infrared spectroscopy and Raman hyperspectral maps treated with hierarchical cluster analysis and averaged spectra with corresponding second derivatives from each cluster. Spectroscopic maps cover both cancerous (red circle) and noncancerous–stroma (green circle) tissue fragments. Adapted with permission from[34]. Citation: Szymoński K, Lipiec E, Sofińska K, Skirlińska-Nosek K, Milian-Ciesielska K, Szpor J, Czaja M, Seweryn S, Wilkosz N, Birarda G, Piccirilli F, Vaccari L, Szymoński M. Spectroscopic screening of pancreatic cancer. Clin Spect 2021; 3: 100016. Copyright© The Authors 2020. Published by ELSEVIER.
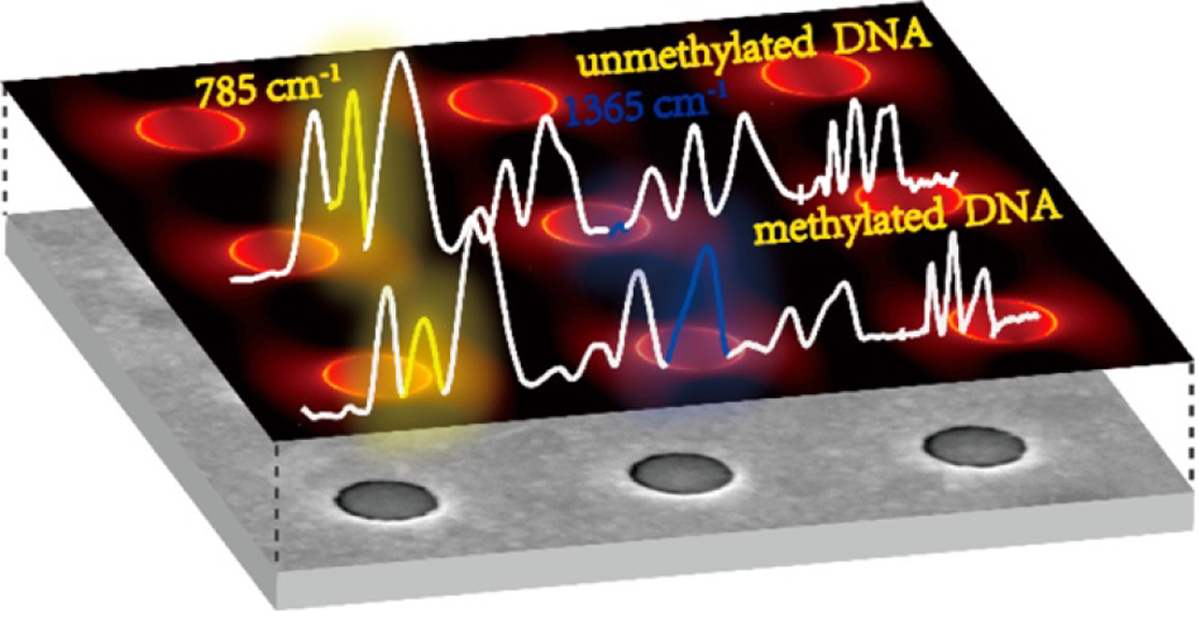
Figure 4 A schematic of a plasmonic gold nanohole array as a surface-enhanced Raman spectroscopy-active substrate used in the detection of DNA methylation[31].
Citation: Luo X, Xing Y, Galvan DD, Zheng E, Wu P, Cai C, Yu Q. Plasmonic Gold Nanohole Array for Surface-Enhanced Raman Scattering Detection of DNA Methylation. ACS Sens 2019; 4: 1534–1542. Copyright© The Authors 2020. Published by American Chemical Society.
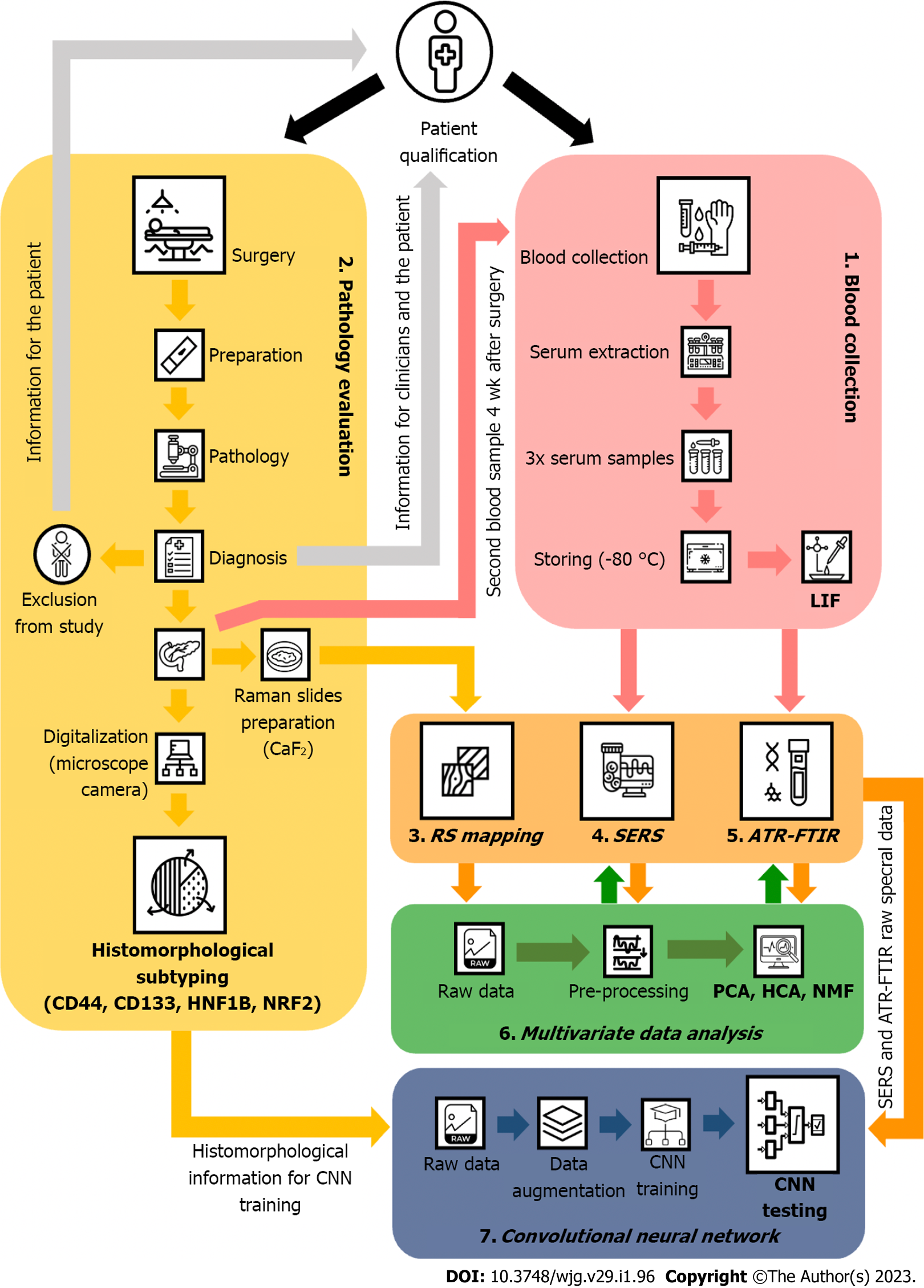
Figure 5 A Designed study regarding the diagnostic potential of vibrational spectroscopic methods used for diagnosing pancreatic cancer by systemically evaluating liquid biopsy samples.
LIF: Leukemia inhibitory factor; RS: Raman spectroscopy; SERS: Surface-enhanced Raman spectroscopy; ATR-FTIR: Attenuated total reflection Fourier transform infrared spectroscopy; CNN: Convolutional neural network; NRF2: Nuclear factor-erythroid 2–related factor 2; HNF1B: Hepatocyte nuclear factor-1B; PCA: Principal component analysis; HCA: Hierarchical clustering analysis; NMF: Non-negative matrix factorization.
- Citation: Szymoński K, Chmura Ł, Lipiec E, Adamek D. Vibrational spectroscopy – are we close to finding a solution for early pancreatic cancer diagnosis? World J Gastroenterol 2023; 29(1): 96-109
- URL: https://www.wjgnet.com/1007-9327/full/v29/i1/96.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i1.96