Copyright
©The Author(s) 2021.
World J Gastroenterol. Jul 21, 2021; 27(27): 4358-4370
Published online Jul 21, 2021. doi: 10.3748/wjg.v27.i27.4358
Published online Jul 21, 2021. doi: 10.3748/wjg.v27.i27.4358
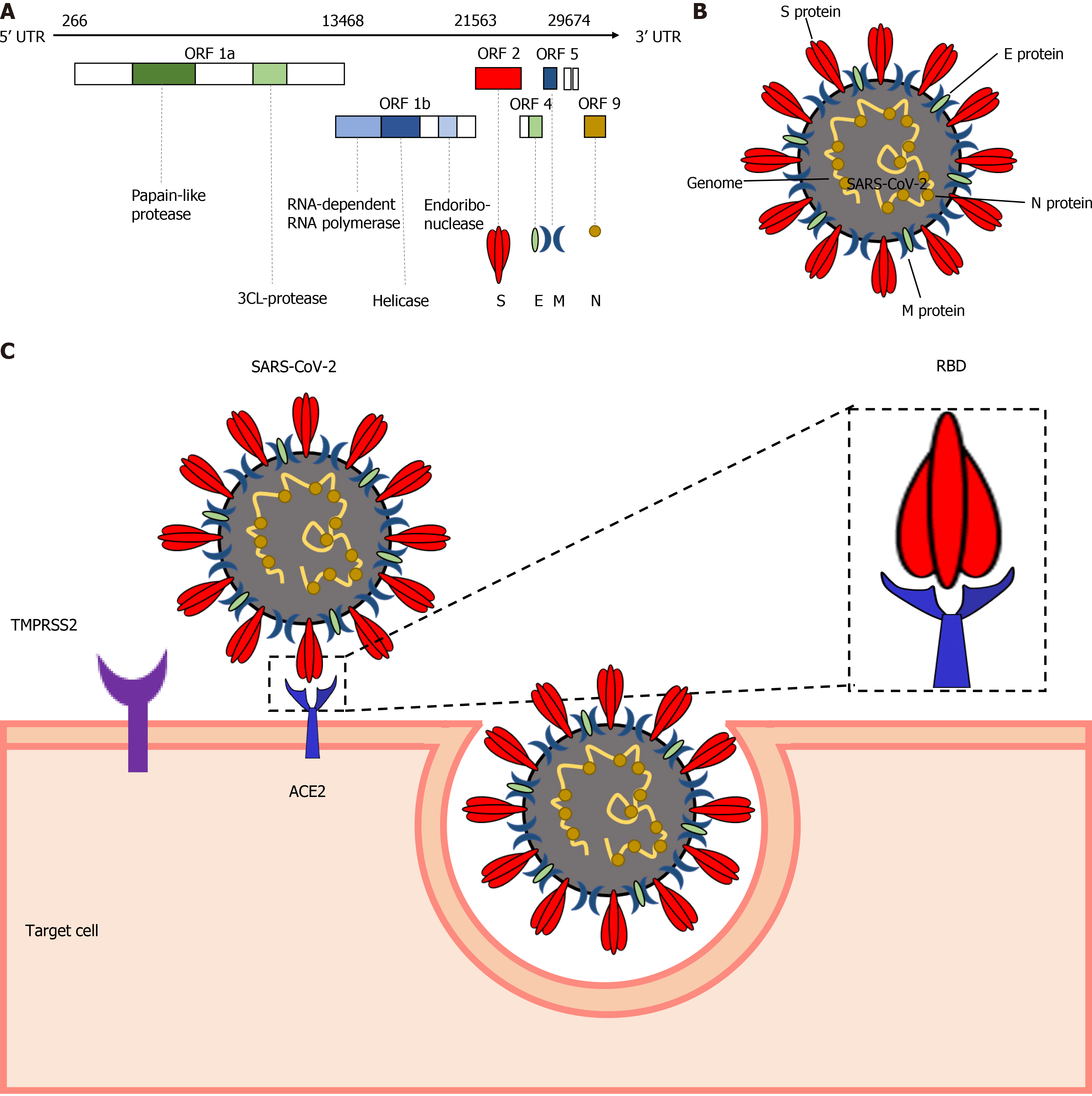
Figure 1 Severe acute respiratory syndrome coronavirus 2 genome and its pathogenesis.
A: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome structure and its encoded proteins; B: Virion structure of SARS-CoV-2 illustrating viral structural proteins including spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N); C: SARS-CoV-2 entry to the target cells by binding the receptor-binding domain of S protein to cellular receptors, such as angiotensin-converting enzyme 2 and transmembrane protease serine 2. SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; RBD: Receptor-binding domain; TMPRSS2: Transmembrane protease serine 2; ACE2: Angiotensin-converting enzyme 2.
Figure 2 Schematic diagram of the two mechanisms of severe acute respiratory syndrome coronavirus 2-induced liver dysfunction.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes direct liver injury through binding to the angiotensin-converting enzyme 2 receptor and transmembrane protease serine 2 expressed on cholangiocytes and hepatocytes. Indirect injury by cytokine storm. T cells are stimulated to secrete large quantities of cytokines including type I interferons, interleukin-6, and tumor necrosis factor-α following SARS-CoV-2 infection, leading to systemic excessive inflammation syndrome. ACE2: Angiotensin-converting enzyme 2; TMPRSS2: Transmembrane protease serine 2; TNF-α: Tumor necrosis factor-α; IL-6: Interleukin-6; IFN: Interferons.
- Citation: Huang YK, Li YJ, Li B, Wang P, Wang QH. Dysregulated liver function in SARS-CoV-2 infection: Current understanding and perspectives. World J Gastroenterol 2021; 27(27): 4358-4370
- URL: https://www.wjgnet.com/1007-9327/full/v27/i27/4358.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i27.4358