Copyright
©The Author(s) 2019.
World J Gastroenterol. Mar 14, 2019; 25(10): 1210-1223
Published online Mar 14, 2019. doi: 10.3748/wjg.v25.i10.1210
Published online Mar 14, 2019. doi: 10.3748/wjg.v25.i10.1210
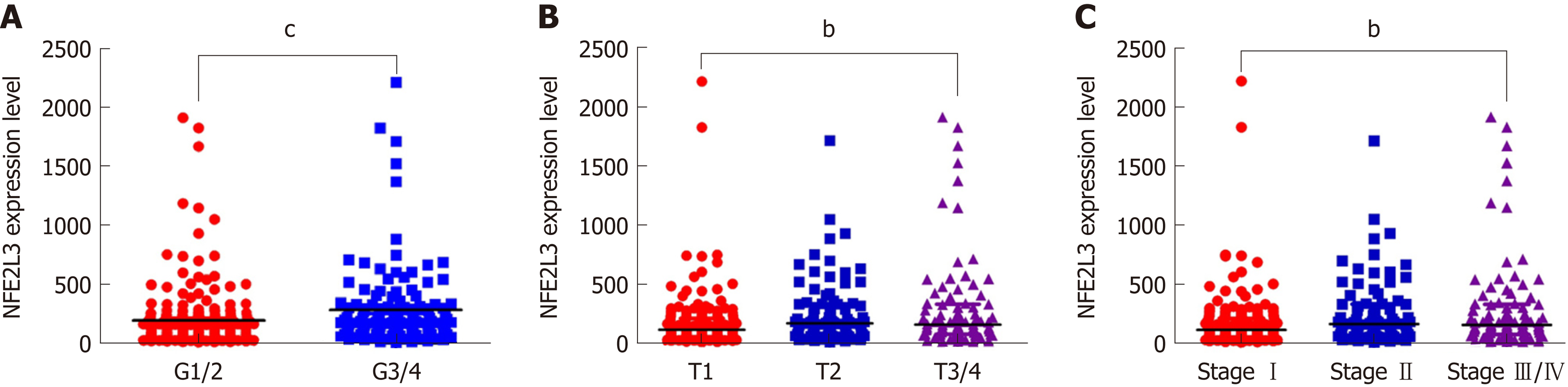
Figure 1 Association of nuclear factor erythroid 2-like 3 expression with clinicopathological features of hepatocellular carcinoma patients in The Cancer Genome Atlas dataset.
A: Nuclear factor erythroid 2-like 3 (NFE2L3) expression in G1/2 (n = 213) and G3/4 (n = 131) hepatocellular carcinoma (HCC) patients; B: NFE2L3 expression in T1 (n = 171), T2 (n = 87) and T3/4 (n = 86) HCC patients; C: NFE2L3 expression in stage I (n = 169), stage II (n = 85), and stage III/IV (n = 90) HCC patients. bP < 0.01, cP < 0.001. NFE2L3: Nuclear factor erythroid 2-like 3; HCC: Hepatocellular carcinoma.
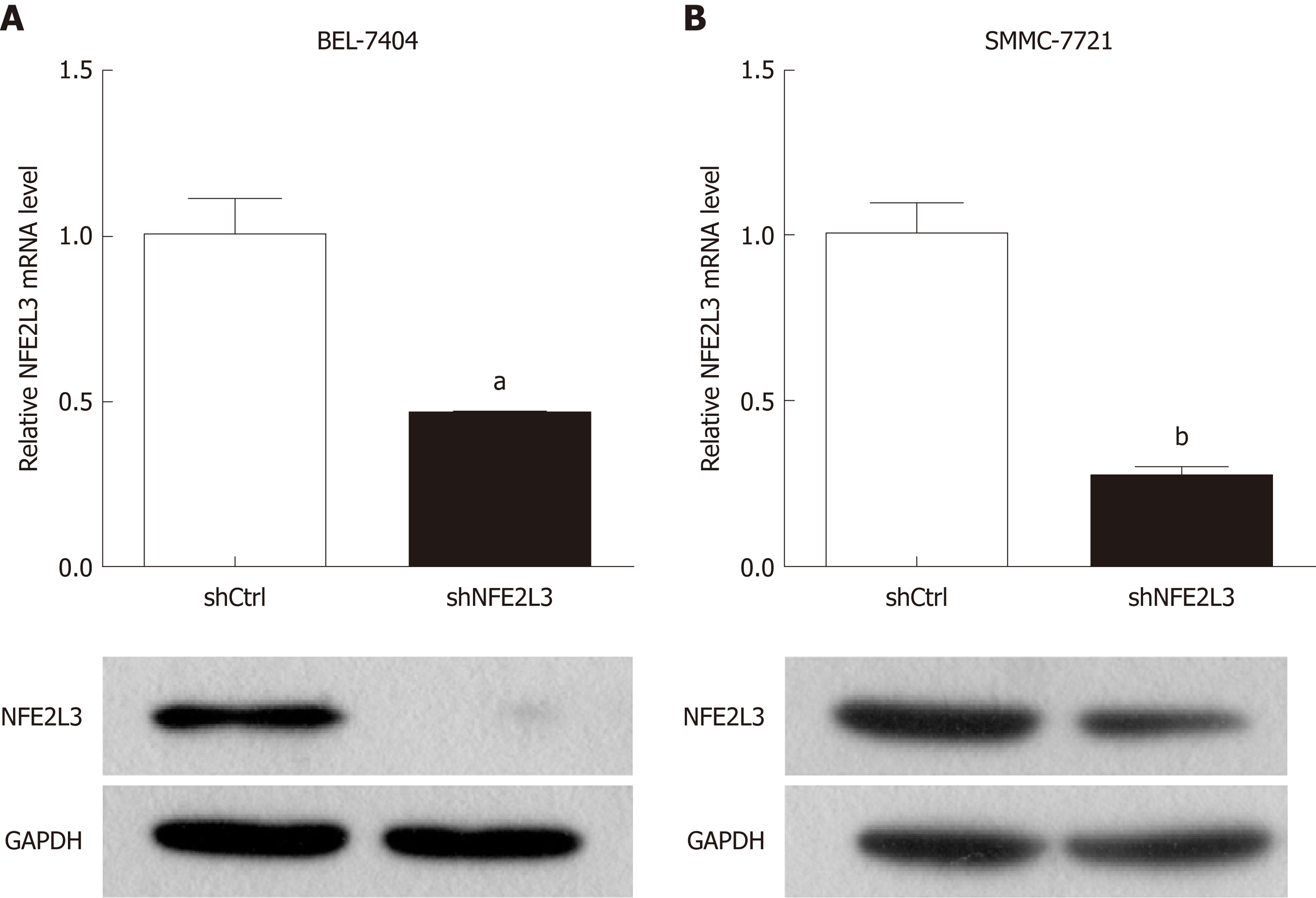
Figure 2 Expression of nuclear factor erythroid 2-like 3 mRNA and protein in hepatocellular carcinoma cell lines.
A: Nuclear factor erythroid 2-like 3 (NFE2L3) mRNA levels in BEL-7404 and SMMC-7721 cells infected with shNFE2L3 (NFE2L3 short hairpin RNA) were measured by qPCR; B: NFE2L3 protein levels in BEL-7404 and SMMC-7721 cells infected with shNFE2L3 were measured by Western blot. Data shown are the mean ± SEM, aP < 0.05, bP < 0.01 vs shCtrl. NFE2L3: Nuclear factor erythroid 2-like 3; shCtrl: Negative control cells; shNFE2L3: Nuclear factor erythroid 2-like 3 short hairpin RNA.
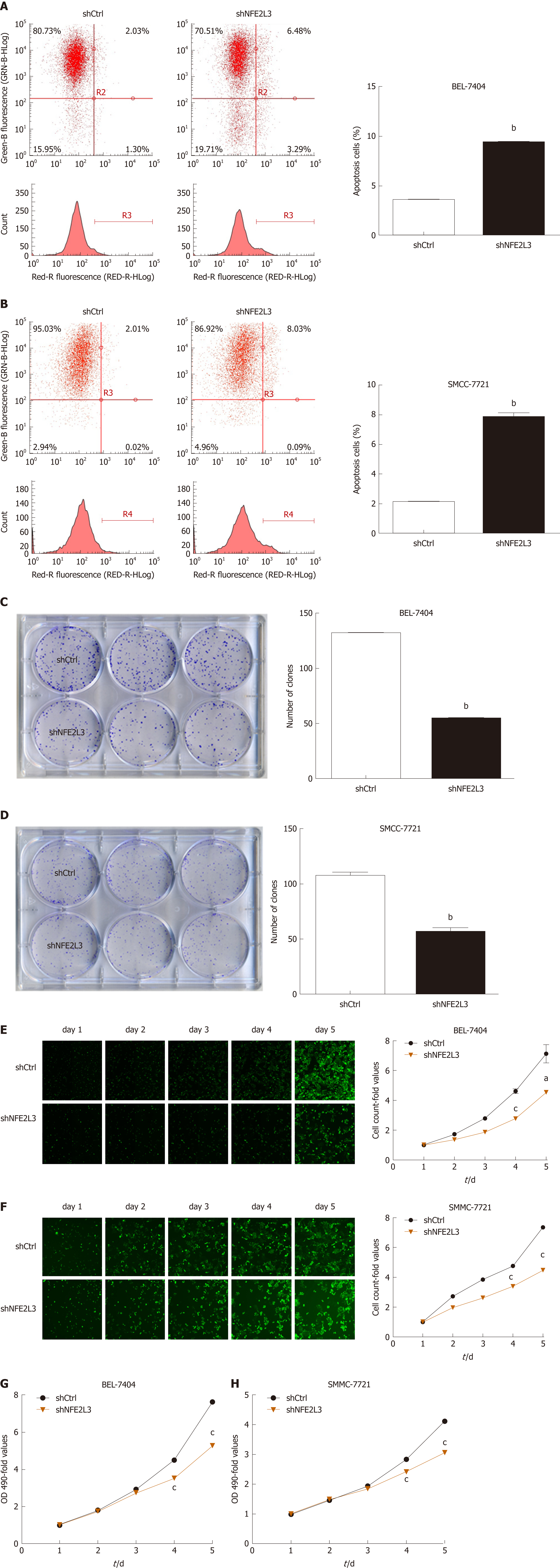
Figure 3 Role of nuclear factor erythroid 2-like 3 in apoptosis, clone formation, and proliferation of hepatocellular carcinoma cells.
A and B: Apoptotic cells were stained with Annexin V-APC and measured using flow cytometry. The abscissa represents red fluorescence (RED-R-HLog), and the ordinate represents green fluorescence (GRN-B-HLog) and cell count; C and D: Cell clones were stained with Giemsa and photographed with a digital camera. The number of clones was accurately calculated and statistically analyzed; E and F: Successfully infected cells were green fluorescent protein positive and cell images were taken with a Celigo cytometer for a continuous 5 d. Cell count-fold represents cell count at each time point relative to the average of day 1; G and H: OD values were measured at 490 nm after the treatment of MTT. Cell growth curves were plotted based on OD 490-fold at different time point. Data shown are the mean ± SEM. Student’s t test was used to analyze significant differences, aP < 0.05, bP < 0.01, cP < 0.001 vs shCtrl. shCtrl: Negative control cells; shNFE2L3: Nuclear factor erythroid 2-like 3 short hairpin RNA.
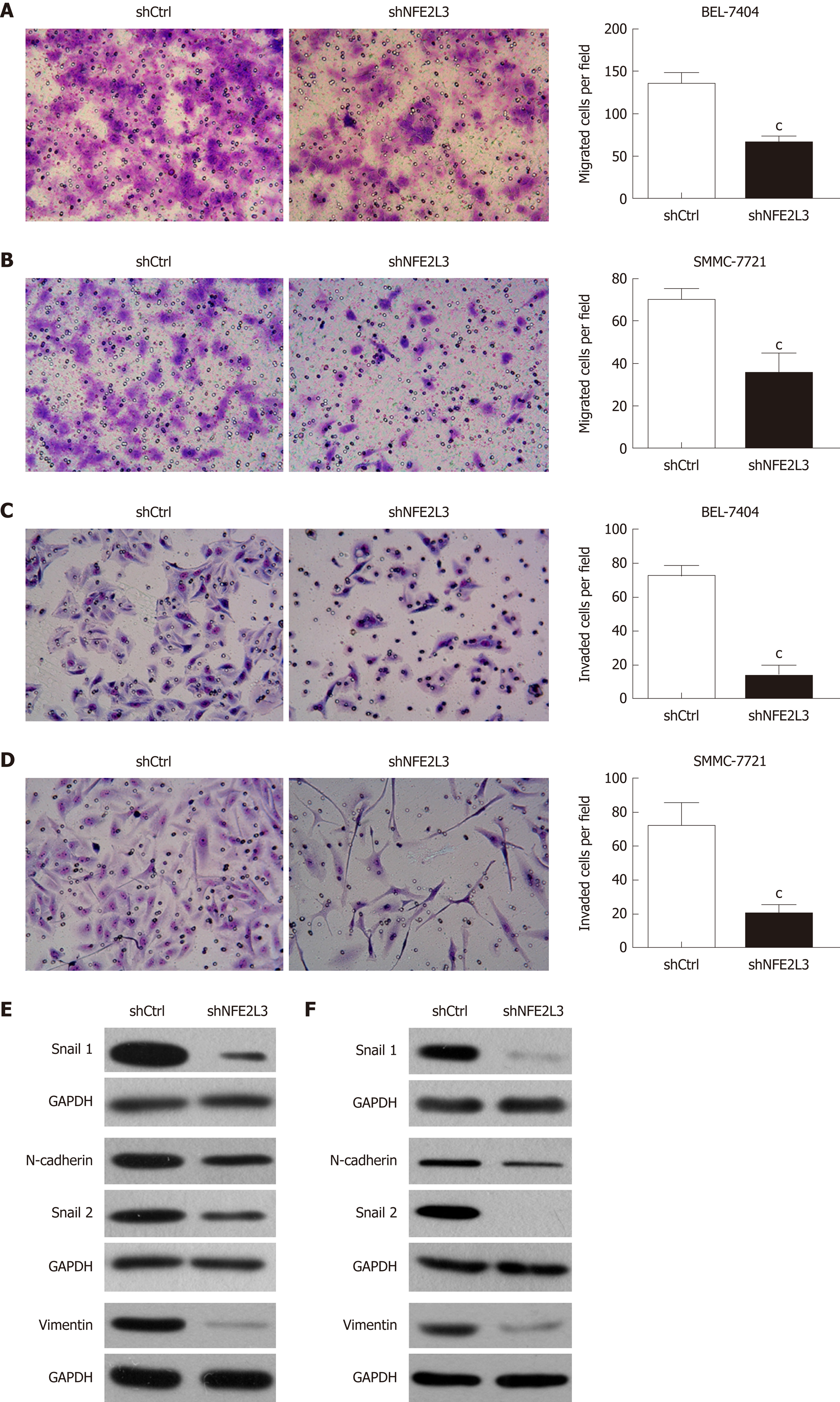
Figure 4 Short hairpin RNA-mediated knockdown of nuclear factor erythroid 2-like 3 suppresses migration, invasion, and epithelial-mesenchymal transition of hepatocellular carcinoma cells.
A and B: Cell migration assay showed that migrated cells of the shNFE2L3 group were less than that of the shCtrl group in BEL-7404 and SMMC-7721 cells; C and D: Cell invasion assay showed that invaded cells of the shNFE2L3 group were less than that of the shCtrl group in BEL-7404 and SMMC-7721 cells. Both cell migration and invasion were measured with transwell assays. Migrated and invaded cells were stained with Giemsa and imaged and counted under a microscope. Scare bar = 150 μm; E and F: Snail 1, N-cadherin, Snail 2, and Vimentin protein levels were analyzed using Western blot. GAPDH was a loading control. Data shown are the mean ± SEM. Student’s t test was used to analyze significant differences, cP < 0.001 vs shCtrl. shCtrl: Negative control cells; shNFE2L3: Nuclear factor erythroid 2-like 3 short hairpin RNA.
- Citation: Yu MM, Feng YH, Zheng L, Zhang J, Luo GH. Short hairpin RNA-mediated knockdown of nuclear factor erythroid 2-like 3 exhibits tumor-suppressing effects in hepatocellular carcinoma cells. World J Gastroenterol 2019; 25(10): 1210-1223
- URL: https://www.wjgnet.com/1007-9327/full/v25/i10/1210.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i10.1210