Copyright
©The Author(s) 2016.
World J Gastroenterol. Apr 7, 2016; 22(13): 3531-3546
Published online Apr 7, 2016. doi: 10.3748/wjg.v22.i13.3531
Published online Apr 7, 2016. doi: 10.3748/wjg.v22.i13.3531
Figure 1 Comparison of the amino acid sequences of the 226AA hepatitis B virus, hepatitis B virus surface antigen between occult hepatitis B virus infections, and controls.
Amino acid substitutions are indicated with one-letter codes. Mutations identified in occult hepatitis B virus infections group and control group are in red and black, respectively. Mutations detected in both groups are in blue. Black dots denote amino acid identity with any of the reference strains of genotype A to H retrieved from GenBank (accession numbers: A, X02763; Ba, D00330; Bc, GQ205440; Bj, AB073858; C1, KM999990; C2, KM999991; D, X02496; E, X75657; F, X69798; G, AF160501; H, AY090454). Genotype-specific substitutions are listed. CON indicates control.
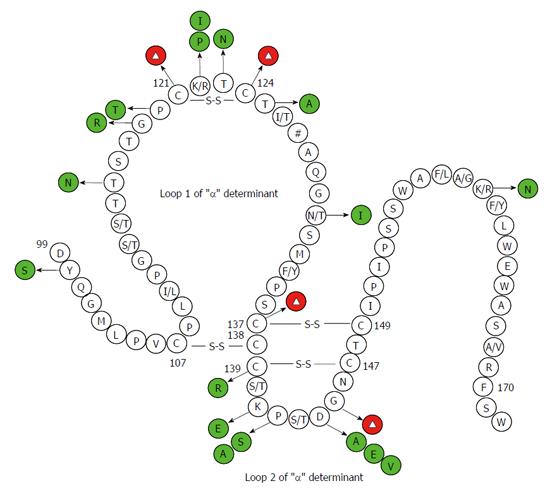
Figure 2 Secondary structure of the major hydrophilic region of the hepatitis B virus, hepatitis B virus surface antigen.
Mutations have been confirmed by functional studies to exert negative effects on antigenicity of hepatitis B virus surface antigen (HBsAg) are shown in green and red circles. Red circles with the Δ symbol denote multiple kinds of amino acid substitutions. These have been substantiated to negatively correlate with HBsAg antigenicity at some sites. The # symbol indicates multiple genotype-specific substitutions. The numbers denote amino acid sites.
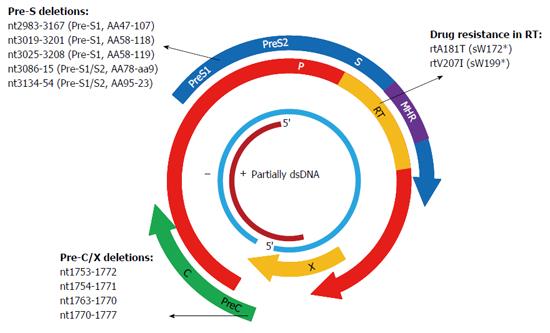
Figure 3 Mutations in X, P, and C genes of hepatitis B virus have been substantiated in association with the occurrence of occult hepatitis B virus infections by in vivo and in vitro studies.
Plus and minus strands are denoted by + and -, respectively.
- Citation: Zhu HL, Li X, Li J, Zhang ZH. Genetic variation of occult hepatitis B virus infection. World J Gastroenterol 2016; 22(13): 3531-3546
- URL: https://www.wjgnet.com/1007-9327/full/v22/i13/3531.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i13.3531