Copyright
©The Author(s) 2015.
World J Gastroenterol. Aug 14, 2015; 21(30): 9093-9102
Published online Aug 14, 2015. doi: 10.3748/wjg.v21.i30.9093
Published online Aug 14, 2015. doi: 10.3748/wjg.v21.i30.9093
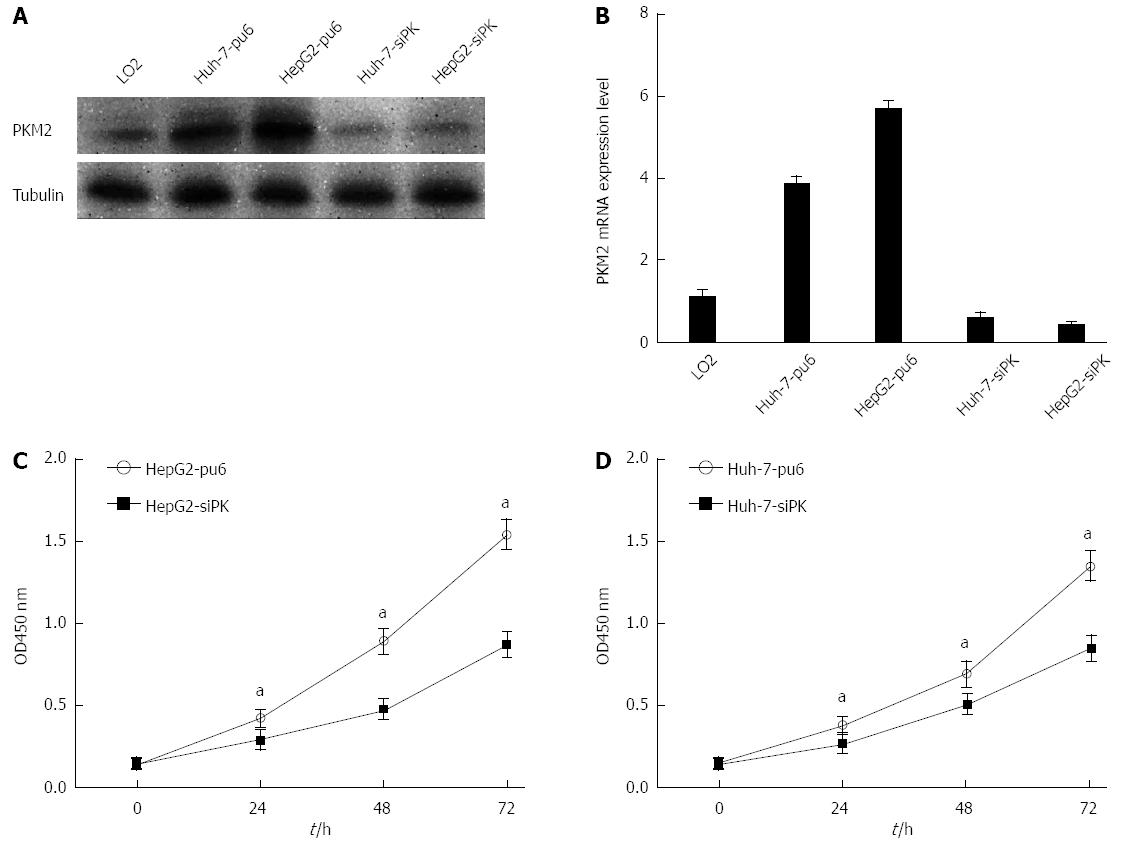
Figure 1 Knockdown of pyruvate kinase M2 isoform suppressed cell proliferation in HepG2 and Huh-7 cells.
A: HepG2 and Huh-7 cells were stably transfected with shRNA directed against pyruvate kinase M2 isoform (PKM2). The specific knockdown of PKM2 was monitored by immunoblot (bottom). Cells stably transfected with pcPUR + U6-siPKM2 are referred to as siPK, and those transfected with pcPUR + U6-siRenilla are referred to as pu6; B: PKM2 expression levels were analyzed by quantitative real-time PCR in stable HepG2 and Huh-7 cells. Error bars represent the mean ± SD of triplicate experiments (aP < 0.05 between groups); C and D: Proliferation of the stably transfected cells. The cell number was determined with the CCK-8 assay, and the relative number of cells is shown.
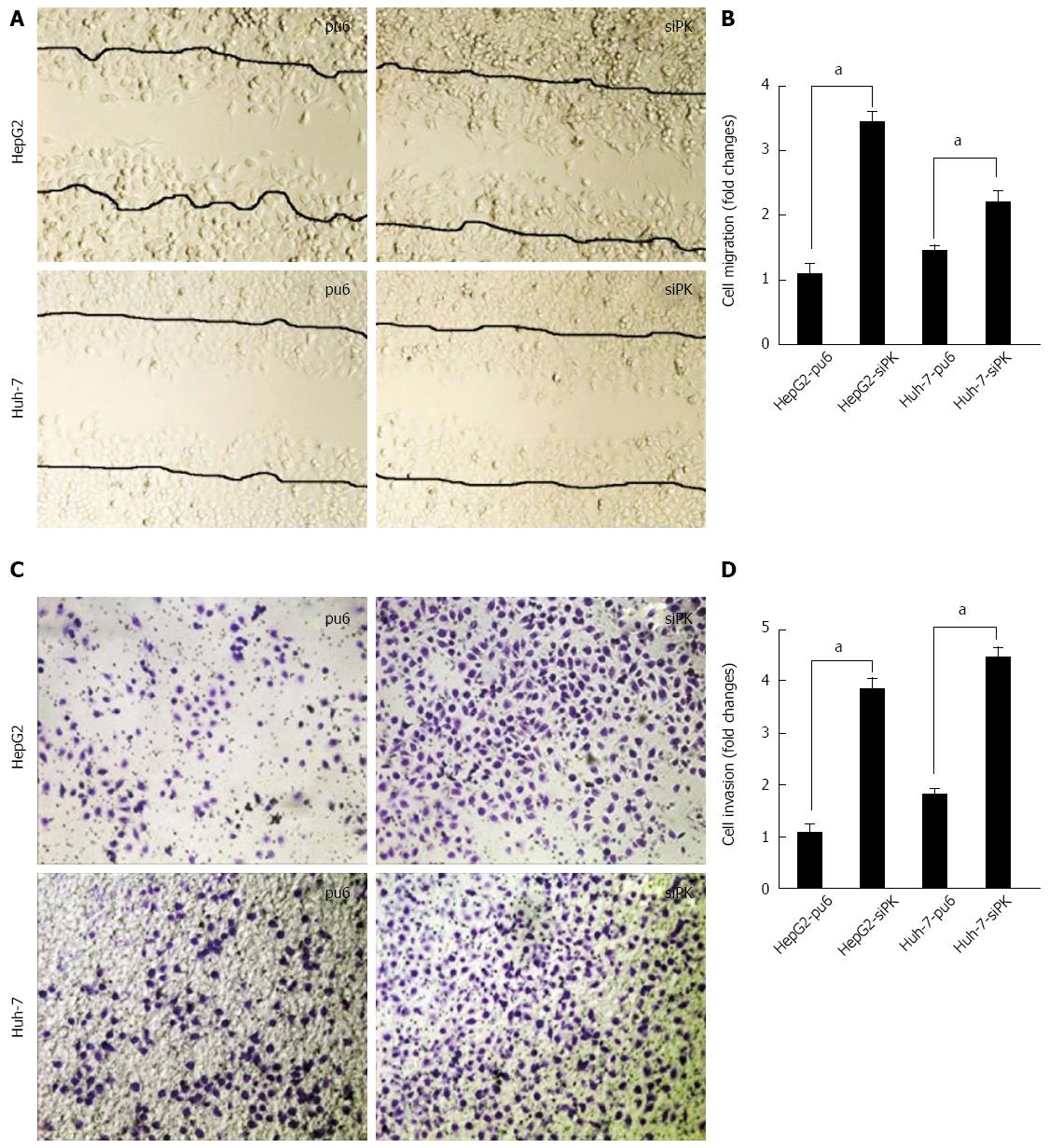
Figure 2 Knockdown of pyruvate kinase M2 isoform promoted the migration and invasion of HepG2 and Huh-7 cells upon epidermal growth factor stimulation.
A: A cross-shaped wound was created in the monolayer, and stably transfected HepG2 and Huh-7 cells were cultured for an additional 24 h with epidermal growth factor (EGF) (50 ng/mL). Representative images of the wounded region are shown; B: The results of the migration assay are also shown as graphs (aP < 0.05); C: The invasion potential of the HepG2 and Huh-7 stable cells was assessed using the BD transwell chamber assay with 50 ng/mL EGF in the lower chamber for 36 h. The cells that migrated to the lower side of the filter were stained, photographed, and counted; D: The data are expressed as the mean ± SD from three independent experiments, aP < 0.05 between groups.
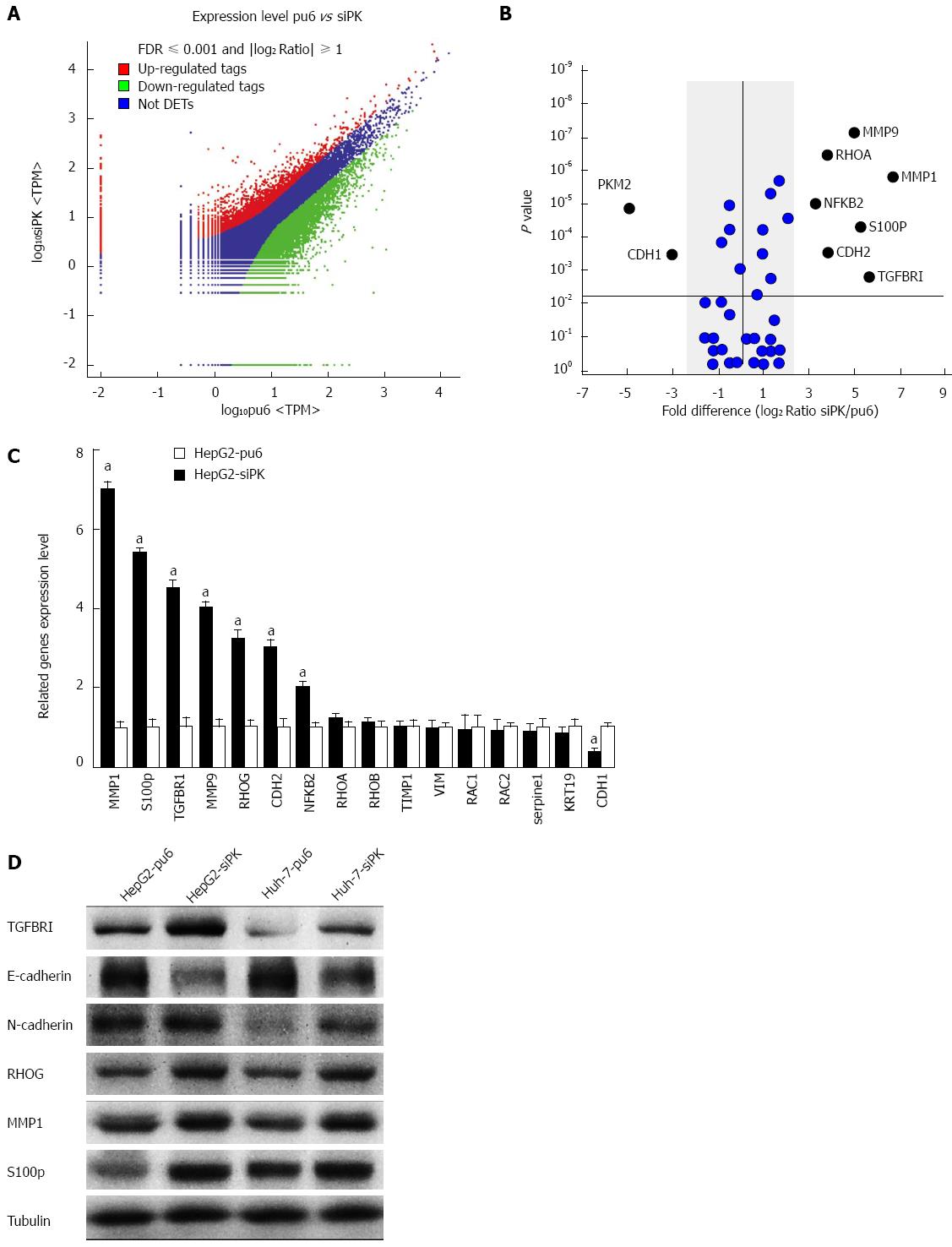
Figure 3 Differential gene expression between HepG2-pu6 and HepG2-siPK cells.
A: The number of statistically significant differentially expressed transcripts identified from the F-test. Color plots show the expression levels of significantly differentially expressed transcripts (P < 0.05 between groups and false discovery rate 0.05) between HepG2-pu6 and HepG2-siPK cells. The upregulated genes are indicated in red, and the downregulated genes are indicated in green. Genes that did not show a statistically significant difference are indicated in blue; B: The graph shows gene expression in relation to cell migration and invasion. Fold differences were calculated based on the log2 ratio of siPK/pu6; C: A comparison of the RT-qPCR and RNA sequence expression analysis; D: The expression levels of genes that were determined to have statistically significant differences in expression were analyzed by immunoblot analysis in stable HepG2 and Huh-7 cells.
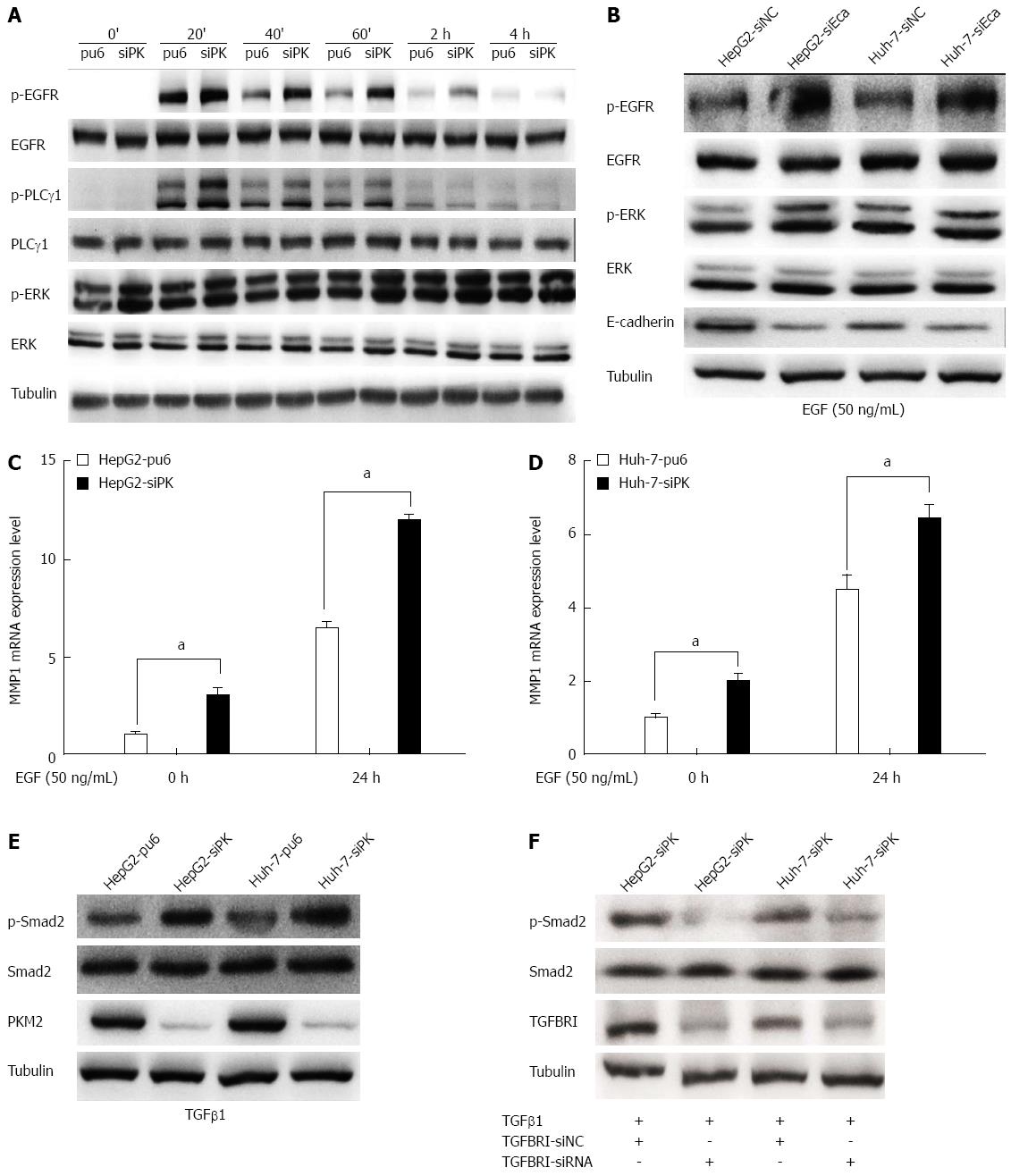
Figure 4 Depletion of pyruvate kinase M2 enhanced the activities of the epidermal growth factor/EGFR and transforming growth factor beta 1/TGFBR downstream signaling pathways.
A: Stable HepG2 and Huh-7 cells were exposed to epidermal growth factor (EGF) (50 ng/mL) for different times. Western blots of the cell lysates are shown. The protein levels of phospho-EGFR (Tyr1068), phospho-PLCγ1 (Tyr783), and phospho-ERK1/2 (Thr202/Tyr204) are shown as indicated; B: The expression of E-cadherin was knocked down in HepG2 and Huh-7 cells by transient transfection of siRNA, and after 48 h, these cells were stimulated with EGF (50 ng/mL). The protein levels of phospho-EGFR (Tyr1068) and phospho-ERK1/2 (Thr202/Tyr204) are shown as indicated; C and D: MMP1 expression levels were analyzed by quantitative real-time PCR in HepG2 and Huh-7 stable cells. Error bars represent the mean ± SD of triplicate experiments (aP < 0.05 between groups); E: Phospho-Smad2 (Ser465/467) protein levels are shown as indicated in stable HepG2 and Huh-7 cells stimulated with TGFβ1 (20 ng/mL); F: The expression of TGFBRI was knocked down in HepG2 and Huh-7 cells by transient transfection of siRNA, and after 48 h, these cells were stimulated with transforming growth factor beta 1(TGFβ1) (20 ng/mL). The protein levels of phospho-Smad2 (Ser465/467) are shown as indicated.
- Citation: Chen YL, Song JJ, Chen XC, Xu W, Zhi Q, Liu YP, Xu HZ, Pan JS, Ren JL, Guleng B. Mechanisms of pyruvate kinase M2 isoform inhibits cell motility in hepatocellular carcinoma cells. World J Gastroenterol 2015; 21(30): 9093-9102
- URL: https://www.wjgnet.com/1007-9327/full/v21/i30/9093.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i30.9093