Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. May 21, 2014; 20(19): 5773-5793
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5773
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5773
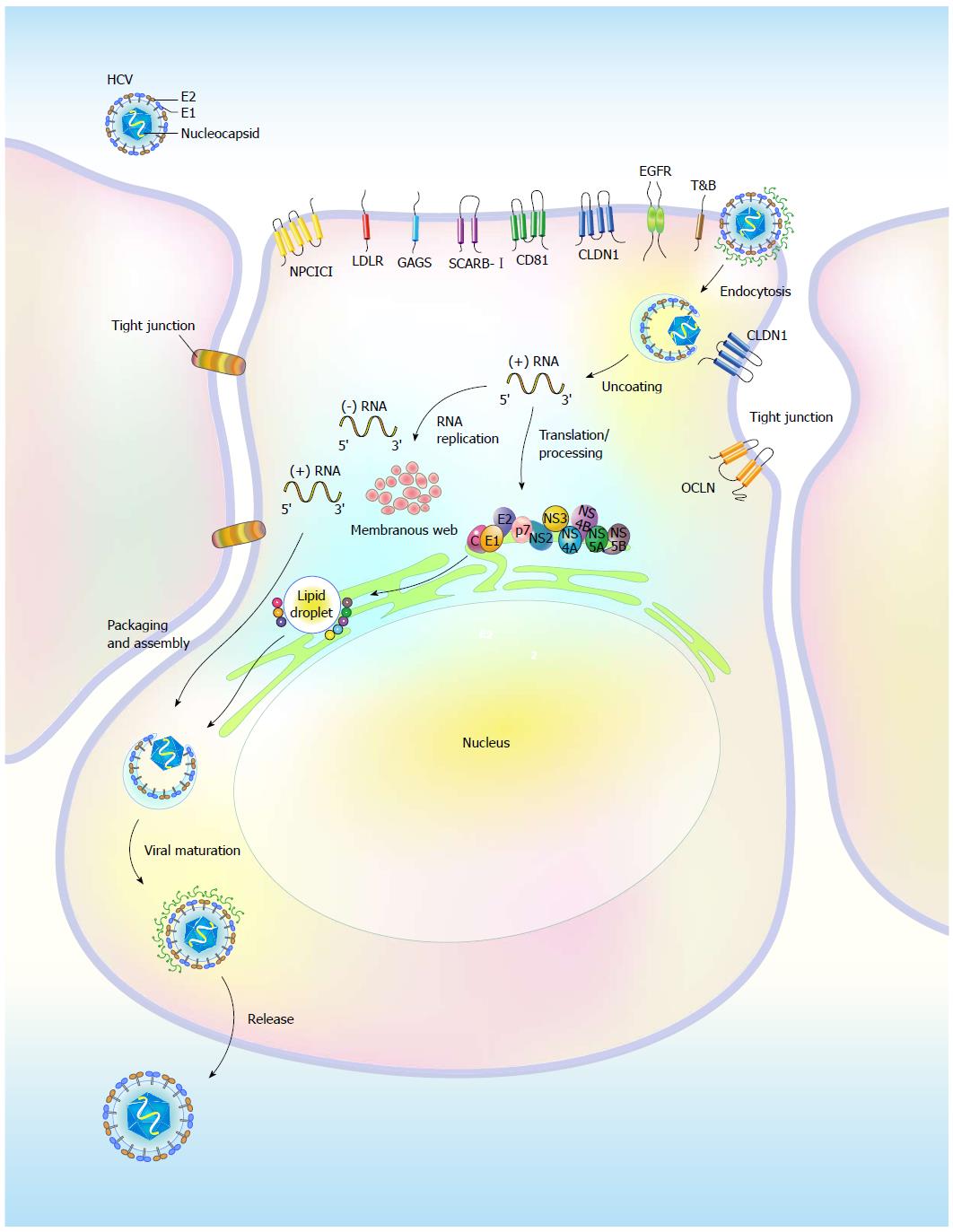
Figure 1 Life cycle of hepatitis C virus.
Hepatitis C virus (HCV) replication involves binding of the virion to the entry molecules on the cell surface of host hepatocytes; endocytosis; viral genome uncoating and translation; polyprotein processing; RNA replication on the ER-associated membrane structure, called the “membranous web”; and virion packaging, assembly, maturation, and release. Glycosaminoglycans (GAG), low-density lipoprotein receptor (LDLR), scavenger receptor class B member I (SCARB-I), two tight junction molecules Claudin 1 (CLDN1) and Occludin (OCLN), epidermal growth factor receptor (EGFR), ephrin receptor A2 (EphA2), Niemann-Pick C1-like L1 (NPC1L1), and transferrin receptor (TfR) constitute the entry receptors. In addition to participating in viral RNA replication, the NS proteins are recruited onto the surface of lipid droplets (LDs) to promote viral RNA replication and the assembly of infectious viral particles, which are composed of the core, E1, and E2 proteins.
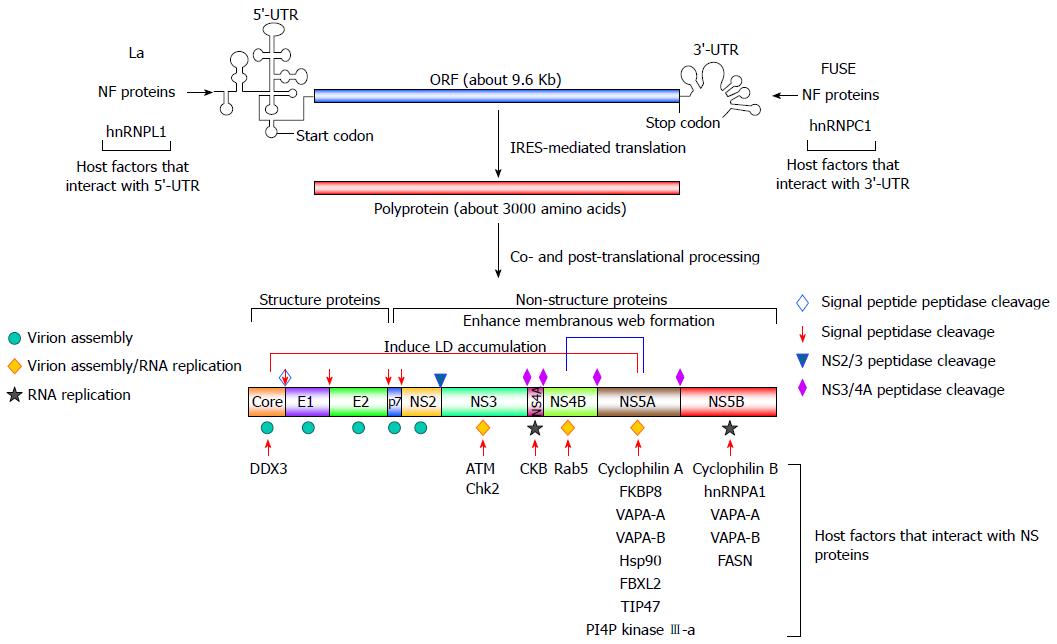
Figure 2 Genome organization of hepatitis C virus and cellular proteins that modulate viral RNA replication.
The positive-sense, single-stranded hepatitis C virus (HCV) genome, containing an open reading frame (ORF) of 9.6 Kb that is flanked by the 5’- and 3’ untranslated regions (UTR), is translated using an internal ribosome entry site (IRES) to a polyprotein, which is processed by cellular and viral proteases to mature into three structural proteins (core, envelope glycoproteins E1 and E2) and seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The role of each viral protein in virus assembly, RNA replication, or both is indicated by the symbol shown on the left. The cellular proteins that modulate HCV replication and their interacting viral counterparts are shown. Host factors that interact with the 5’-UTR and 3’-UTR, as indicated, regulate IRES-mediated translation and viral RNA replication, respectively. Membrane-associated proteins, as shown, modulate HCV replication through interactions with NS5A and NS5B. Other cellular factors, such as DDX3, ATM and Chk2, CKB, and Rab5, promote the replication of HCV viral RNA via interacting with core, NS3, NS4A, and NS4B, respectively.
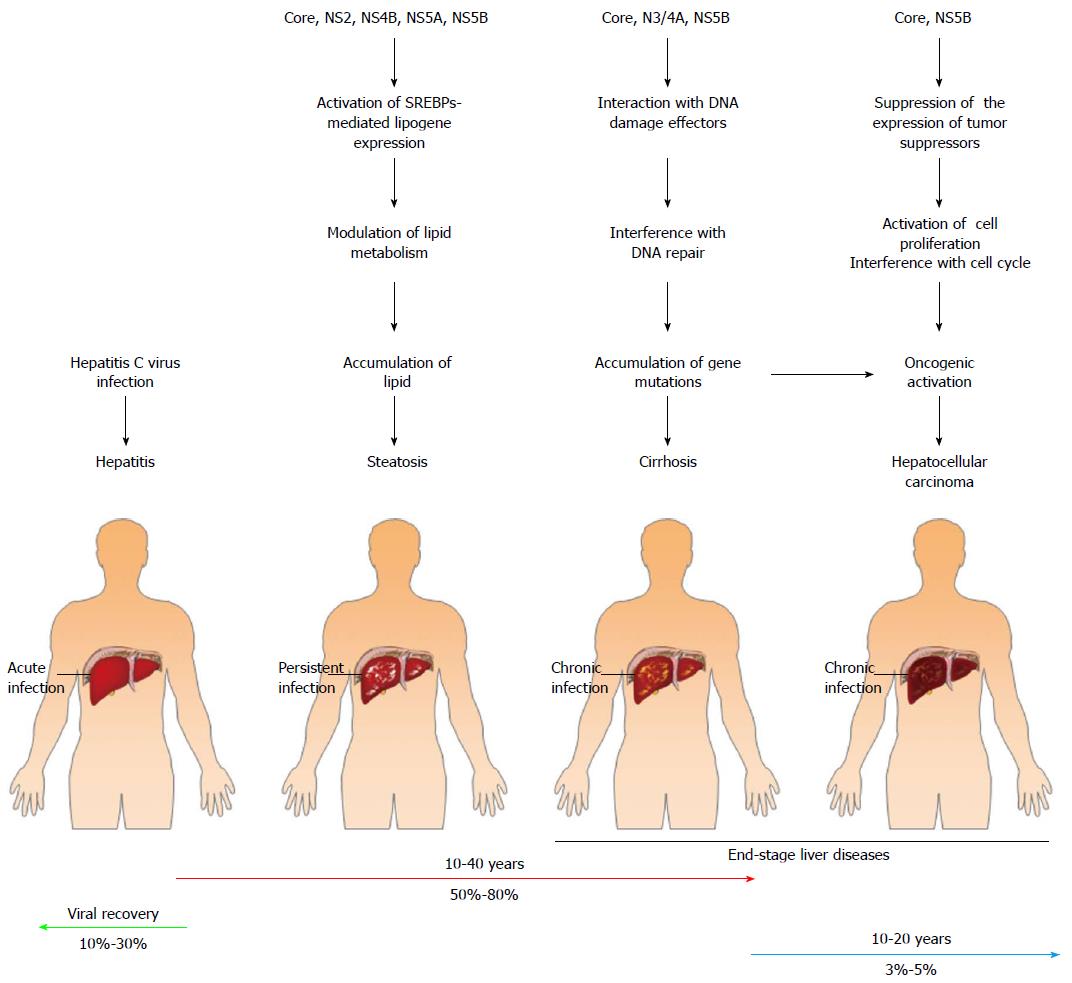
Figure 3 Contribution of viral proteins to the pathogenic changes leading to hepatitis C virus-related liver diseases.
After acute hepatitis C virus (HCV) infection, approximately 10%-30% of cases will spontaneously recover, whereas the majority of HCV infection cases (approximately 50%-80% of infected cases) become persistently infected. Chronically HCV-infected hepatocytes in the livers of these patients progress into hepatosteatosis and liver cirrhosis. Ultimately, some (approximately 3%-5%) of the chronically infected individuals develop end-stage liver disease, i.e., hepatocellular carcinoma. The potential contributions of the viral proteins to the development of HCV-associated liver diseases are shown. The HCV core protein and the NS2, NS4, NS5A, and NS5B proteins can promote hepatic steatosis by mediating the clustering of LDs and regulating the SREBP-mediated transactivation of lipogenes, respectively. The binding of core and NS5B to tumor suppressors downregulates the expression of these tumor suppressors, thus activating cell proliferation and interfering with cell cycle progression. Moreover, HCV core, NS3/4A, and NS5B can bind to DNA damage response-associated effectors, thereby inhibiting the DNA repair mechanism and inducing genome instability. Deregulated cell growth, cell cycle progression, and genome instability may, in turn, trigger oncogenic activation in infected cells, which finally leads to the development of hepatocellular carcinoma.
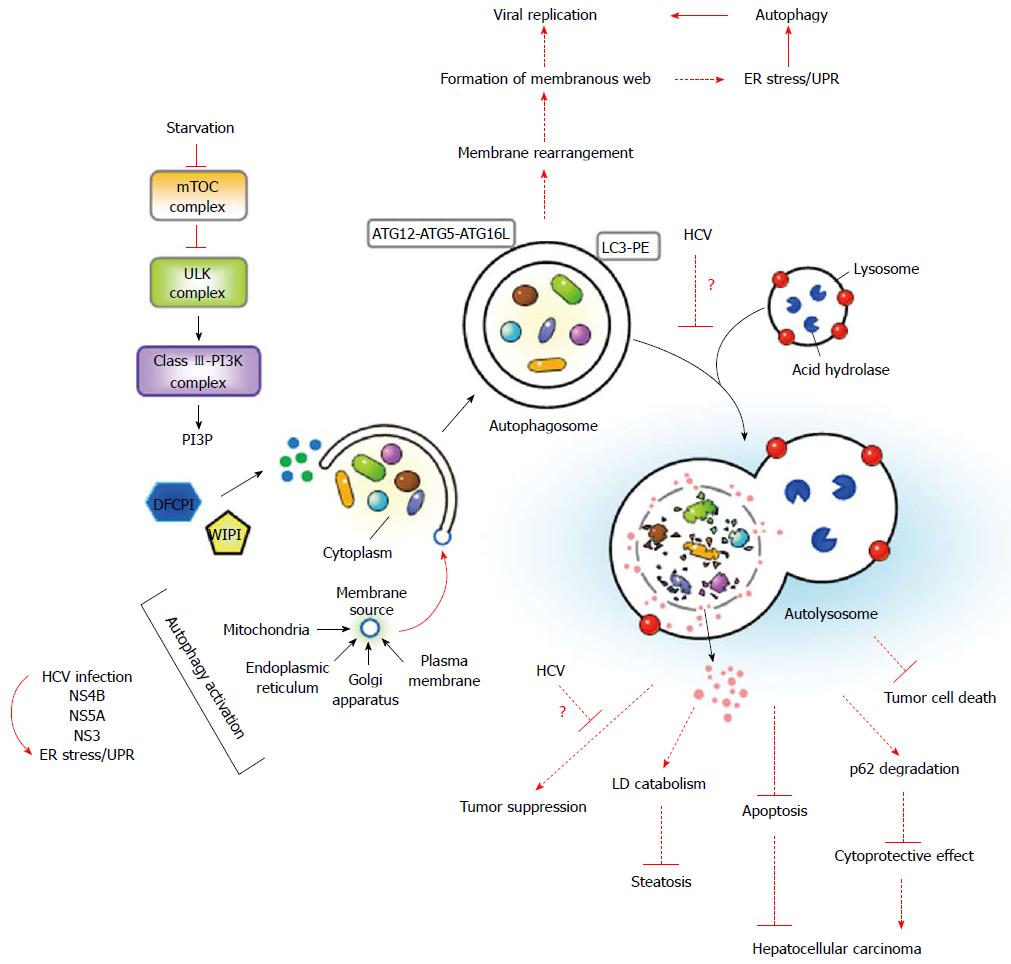
Figure 4 Sequential and coordinated events of autophagy and their impact on viral replication and hepatitis C virus-associated liver diseases.
When cells undergo nutrient starvation, the activity of the mammalian target of rapamycin (mTOR) complex is inhibited, resulting in dephosphorylation, activation, and translocation of the unc-51 like-kinase (ULK) complex to the ER, where the ULK complex activates the class III phosphatidylinositol-3-OH kinase (class III-PI3K) complex to generate PtdIn(3)P (PI3P). PI3P, in turn, recruits double-FYVE-containing protein 1 (DFCP1) and WD-repeat domain PI3P-interacting (WIPI) protein into the isolation membrane, which may originate from the ER, mitochondria, plasma membrane, or Golgi apparatus. Two ubiquitin-like conjugation systems, the ATG12-ATG5-ATG16L and LC3-PE conjugation cascades, coordinate the elongation and enclosure of the autophagosome. Finally, the autophagosome fuses with a lysosome to degrade the sequestrated cytoplasmic components. Hepatitis C virus (HCV) infection, which activates ER stress/the unfolded protein response (UPR), as marked by a curved arrow, and ectopic expression of NS4B, NS5A, and NS3 are known to activate autophagy. The dashed lines indicate the potential implications of autophagy in HCV replication, HCV-related steatosis, and hepatocellular carcinoma. Activated autophagy may contribute to the rearrangement of the membrane as a resource of the membranous web for viral RNA replication. It is surmised that the HCV-induced blockade in autolysosome formation, as shown by a “?” during fusion of the autophagosome with a lysosome, may disrupt the effect of autophagy on LD and p62 degradation, thus contributing to the development of steatosis and hepatocellular carcinoma. On the other hand, interference with the effects of autophagy on tumor suppression by HCV, as indicted by a “?”, or the inhibitory effects of autophagy on the apoptotic signaling and killing of tumor cells in HCV-infected patients may also enhance the progression to hepatocellular carcinoma.
- Citation: Ke PY, Chen SSL. Autophagy in hepatitis C virus-host interactions: Potential roles and therapeutic targets for liver-associated diseases. World J Gastroenterol 2014; 20(19): 5773-5793
- URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5773.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5773