Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. May 21, 2014; 20(19): 5666-5671
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5666
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5666
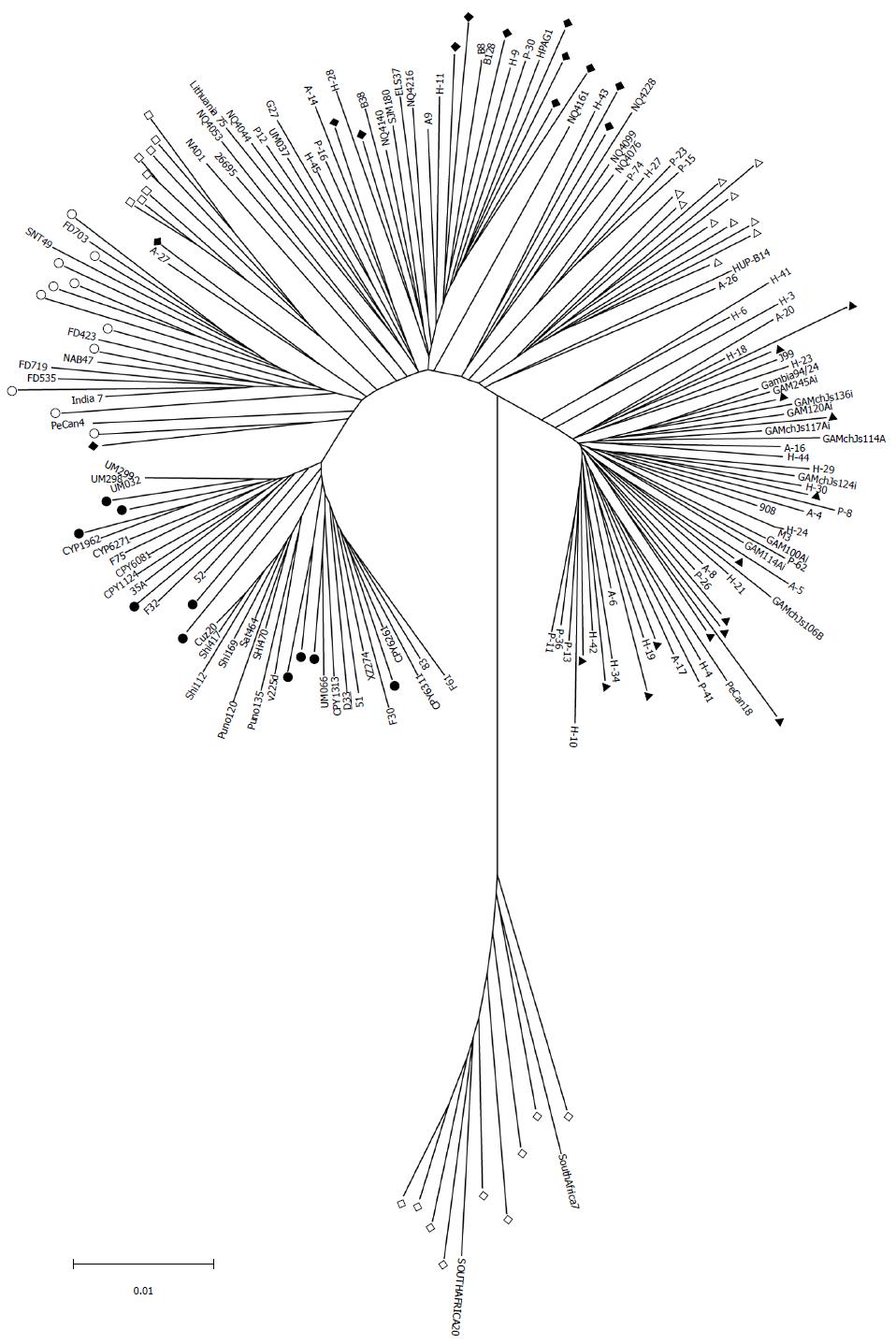
Figure 1 Neighbor-joining tree of 118 sequenced Helicobacter pylori strains and 77 strains from multilocus sequence typing database based on the phylogenetic analysis of seven house-keeping genes (atpA, efp, mutY, ppa, trpC, ureI and yphC).
The population to which those 77 strains belong is known. The tree was constructed with Mega 5.0 software. Scale bar indicates substitutions per nucleic acid residue (change/nucleotide site). Classification of population/ subpopulation was as previously described[6,7].
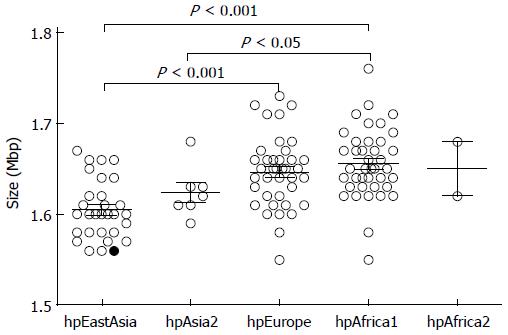
Figure 2 Genome size of strains from different Helicobacter pylori populations.
Single factor analysis of variance was used for the statistical analysis. The average genome size was significantly different between Helicobacter pylori populations.
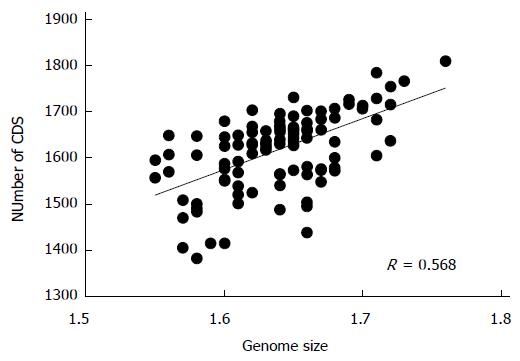
Figure 3 Correlation of the genome size of Helicobacter pylori with the number of protein coding genes.
CDS: Coding sequence.
-
Citation: Dong QJ, Wang LL, Tian ZB, Yu XJ, Jia SJ, Xuan SY. Reduced genome size of
Helicobacter pylori originating from East Asia. World J Gastroenterol 2014; 20(19): 5666-5671 - URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5666.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5666