Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Dec 7, 2013; 19(45): 8211-8218
Published online Dec 7, 2013. doi: 10.3748/wjg.v19.i45.8211
Published online Dec 7, 2013. doi: 10.3748/wjg.v19.i45.8211
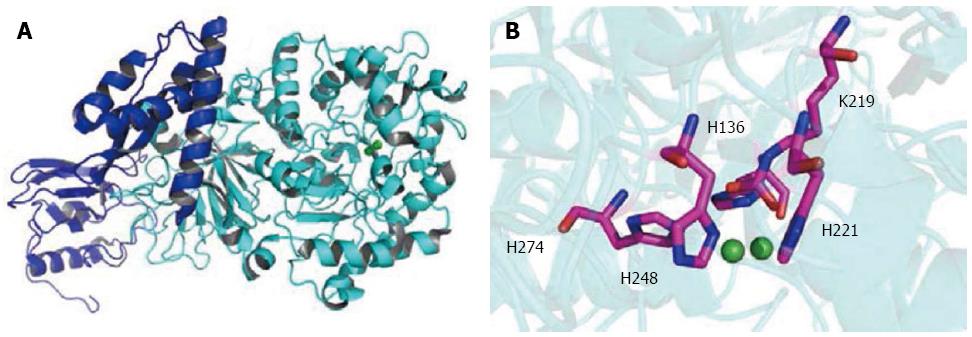
Figure 1 Structure of Helicobacter pylori urease.
A: The urease enzyme subunit UreAB (UreA, blue; UreB, cyan; PDB code: 1E9Z); B: The active sites of H. pylori urease with the side chains of the enzyme involved in the chelation of the catalytic di-nickel center shown (Ni, green; O, red; N, blue; C, pink). H. pylori: Helicobacter pylori.
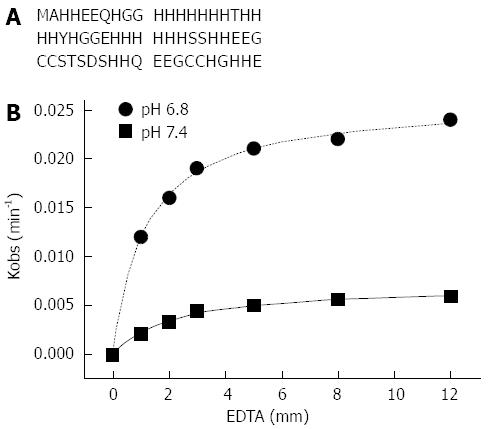
Figure 2 Amino acid sequence of Hpn (A) and the apparent rate of nickel release vs concentrations of ethylenediaminetetraacetic acid at pH 6.
8 and 7.4 and the best nonlinear fit of the data (B). EDTA: Ethylenediaminetetraacetic acid.
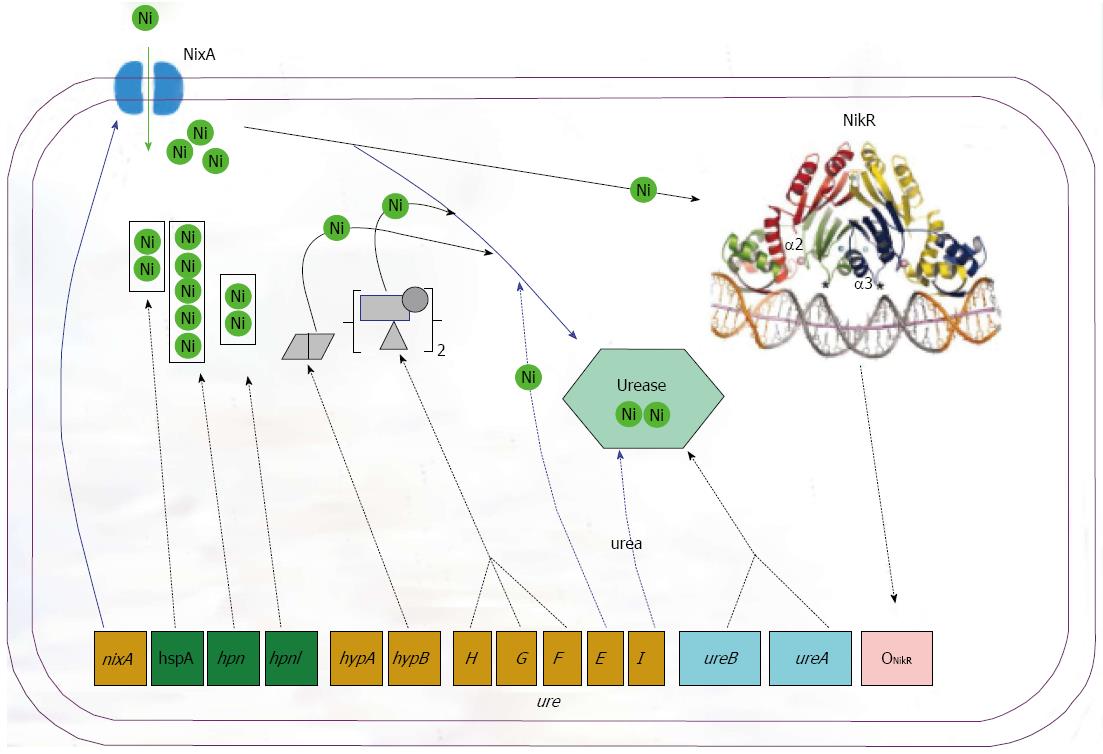
Figure 3 Complex network controlling urease synthesis and activity in Helicobacter pylori.
The different levels of control comprise (1) expression of the UreAB structural subunits fine-tuned by acidity and the nickel-dependent transcriptional regulator NikR; (2) nickel uptake into cells via NixA importer; (3) nickel storage in histidine-rich proteins such as Hpn, Hpnl and HspA; (4) nickel incorporation into urease as mediated by accessory proteins UreEFGH and HypAB; and (5) urea substrate entry via UreI. H. pylori: Helicobacter pylori.
-
Citation: Ge RG, Wang DX, Hao MC, Sun XS. Nickel trafficking system responsible for urease maturation in
Helicobacter pylori . World J Gastroenterol 2013; 19(45): 8211-8218 - URL: https://www.wjgnet.com/1007-9327/full/v19/i45/8211.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i45.8211