Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Jul 28, 2013; 19(28): 4495-4503
Published online Jul 28, 2013. doi: 10.3748/wjg.v19.i28.4495
Published online Jul 28, 2013. doi: 10.3748/wjg.v19.i28.4495
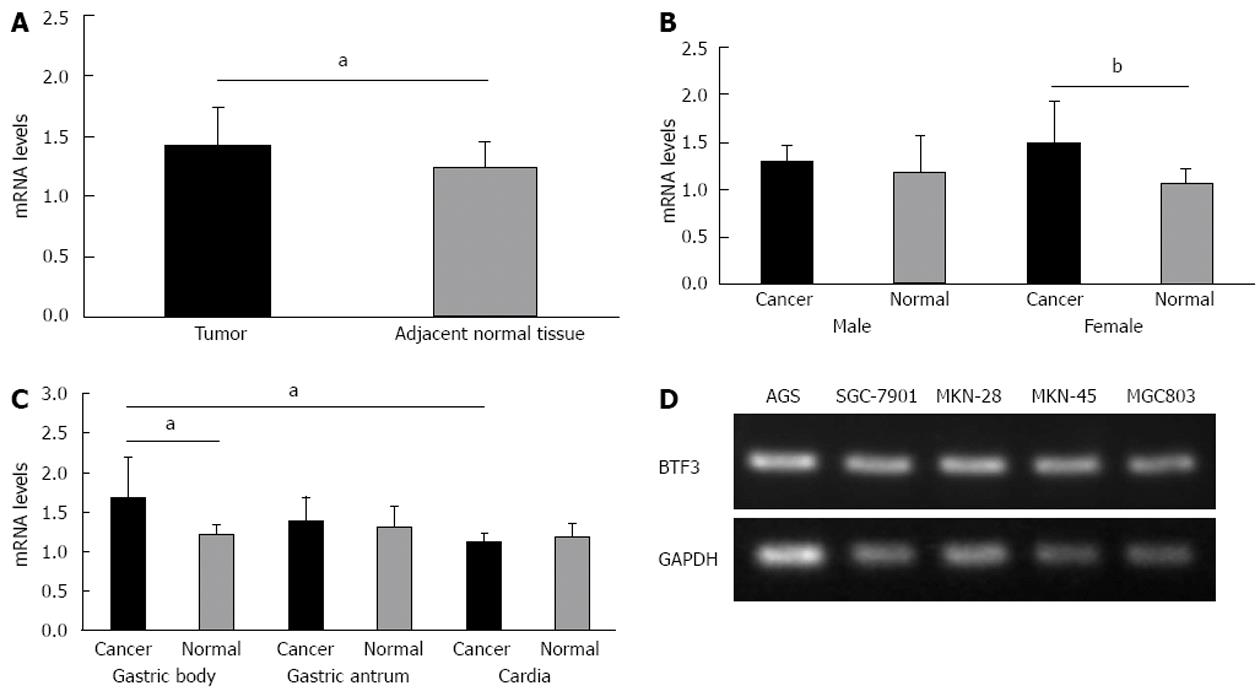
Figure 1 Level of basic transcription factor 3 expression in gastric cancer and tumor cell lines.
A: Bar charts represent the mRNA levels of basic transcription factor 3 (BTF3) in gastric tumor and adjacent normal tissue measured by quantitative real-time polymerase chain reaction (PCR); B: BTF3 mRNA expression data from (A) further classified by gender; C: BTF3 expression pattern among gastric body, gastric antrum and cardia tumors; D: The expression of BTF3 mRNA was detected by quantitative real-time PCR among different cell lines. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene. Black bars represent tumors and gray bars normal tissues. The difference in BTF3 expression, aP < 0.05, bP < 0.01 between groups was assessed by unpaired t test and considered significantly different.
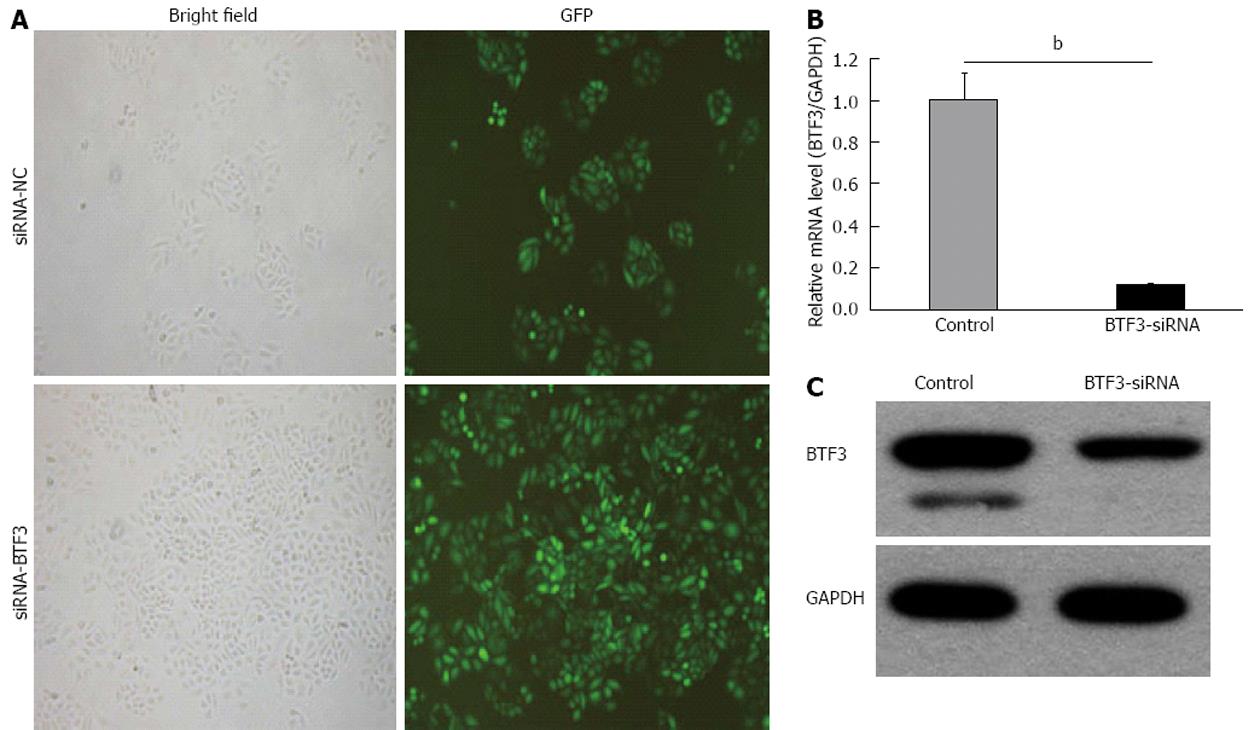
Figure 2 Silencing of basic transcription factor 3 using small interfering RNA.
A: Transfection rates of small interfering RNA (siRNA)-normal control (NC)/siRNA-basic transcription factor 3 (BTF3)-green fluorescent protein (GFP) vectors in human SGC7901 cells, estimated by bright field and fluorescence microscope; B: Bar charts represent quantitative real-time polymerase chain reaction data of BTF3 mRNA levels in control and BTF3-siRNA transfected SGC7901 cells (bP < 0.01 between groups, unpaired t test); C: Immuno-blotting analysis using BTF3-specific antibodies to detect BTF3 protein expressions in control and BTF3-siRNA transfected SGC7901 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control.
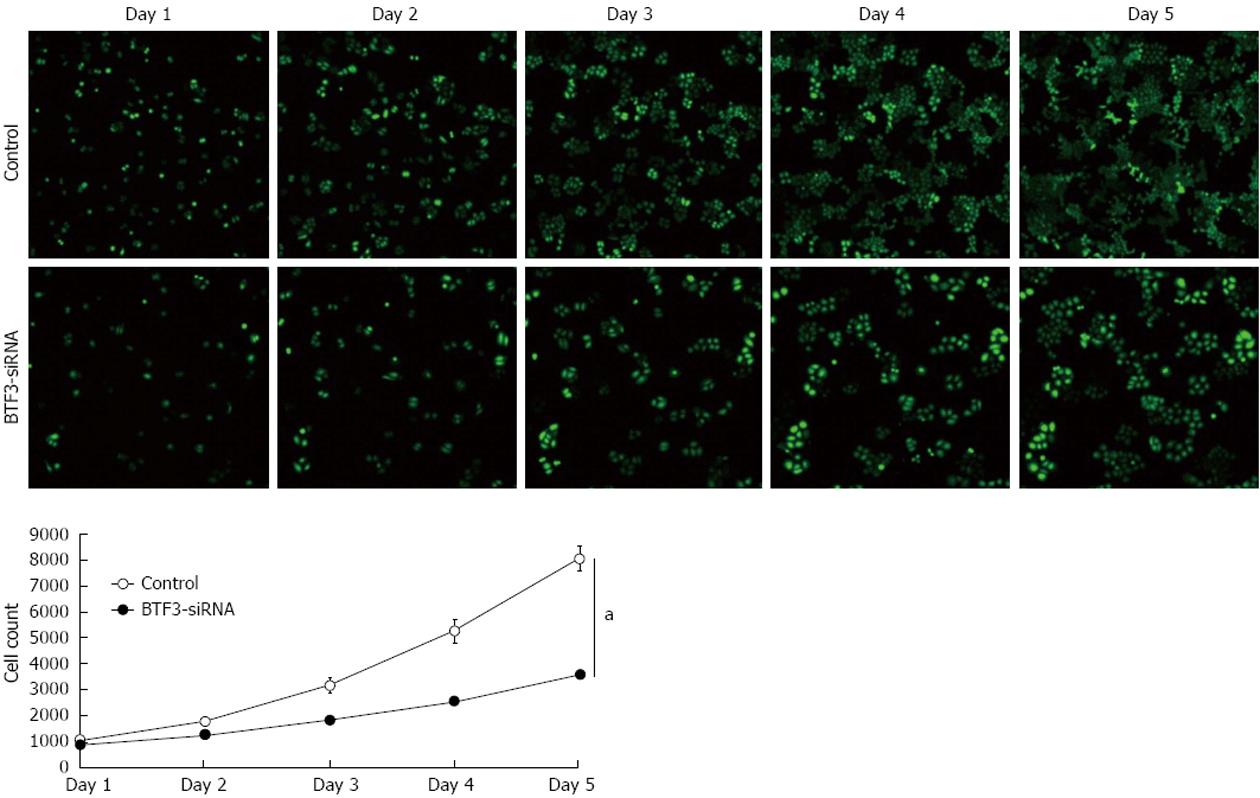
Figure 3 Cell proliferation assay.
Fluorescent images of control and small interfering RNA (siRNA)-basic transcription factor 3 (BTF3) transfected cell proliferation assays. Cells were counted based on the green fluorescent protein signals during 5 d. BTF3-siRNA chart of the cell proliferation assay using two-way analysis of variance. aP < 0.05 between groups.
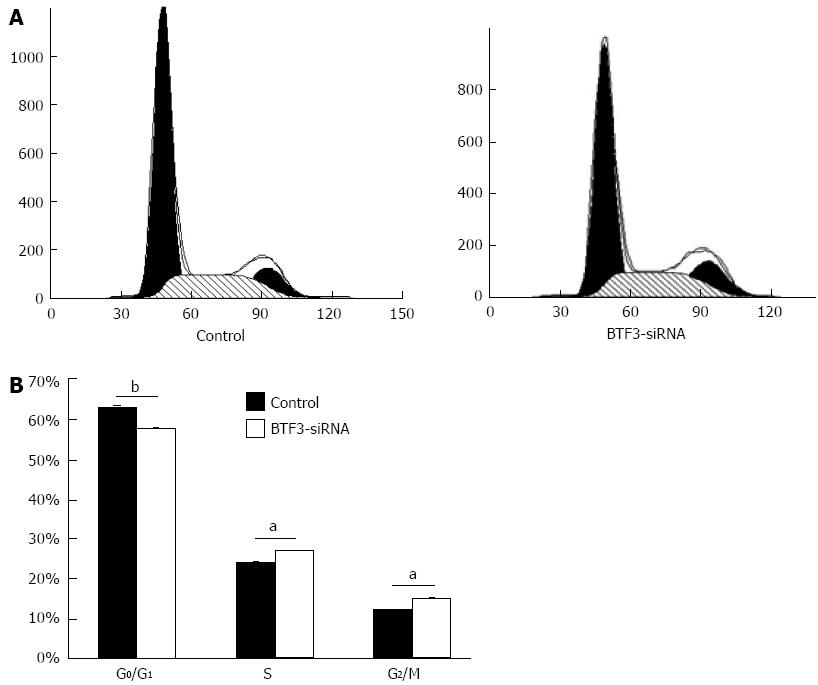
Figure 4 Flow cytometric assays.
A: Cell-cycle stages of control and basic transcription factor 3 (BTF3)-small interfering RNA (siRNA) transfected cells were analyzed by flow cytometry. Data are presented as a histogram, with cell number (y-axis) plotted against DNA content (x-axis); B: Cells arrested at different cell-cycle stages were plotted as bar charts. aP < 0.05, bP < 0.01 between groups was assessed by unpaired t test and comparisons were considered significantly different.
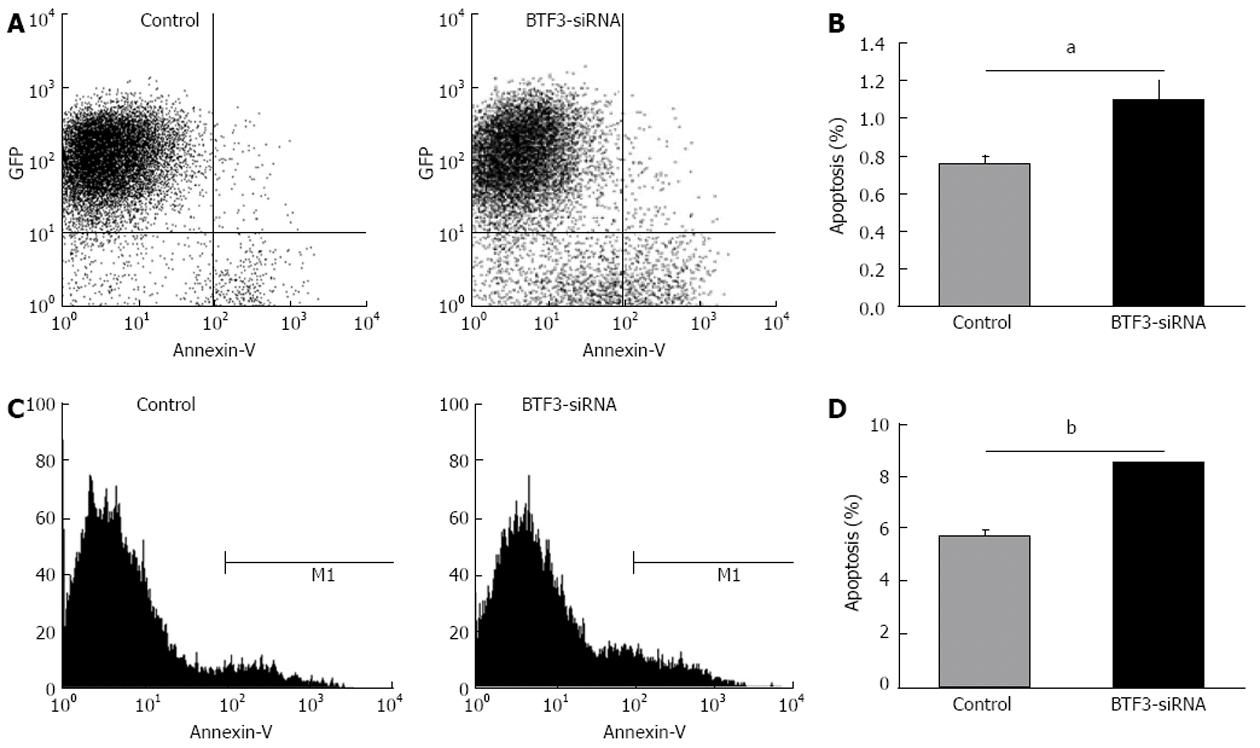
Figure 5 Apoptosis analyses by flow cytometry.
A and C: Flow cytometric analysis of control and basic transcription factor 3 (BTF3)-small interfering RNA (siRNA) transfected cells; B and D: Apoptosis rates plotted as bar charts. aP < 0.05, bP < 0.01 between groups. GFP: Green fluorescent protein.
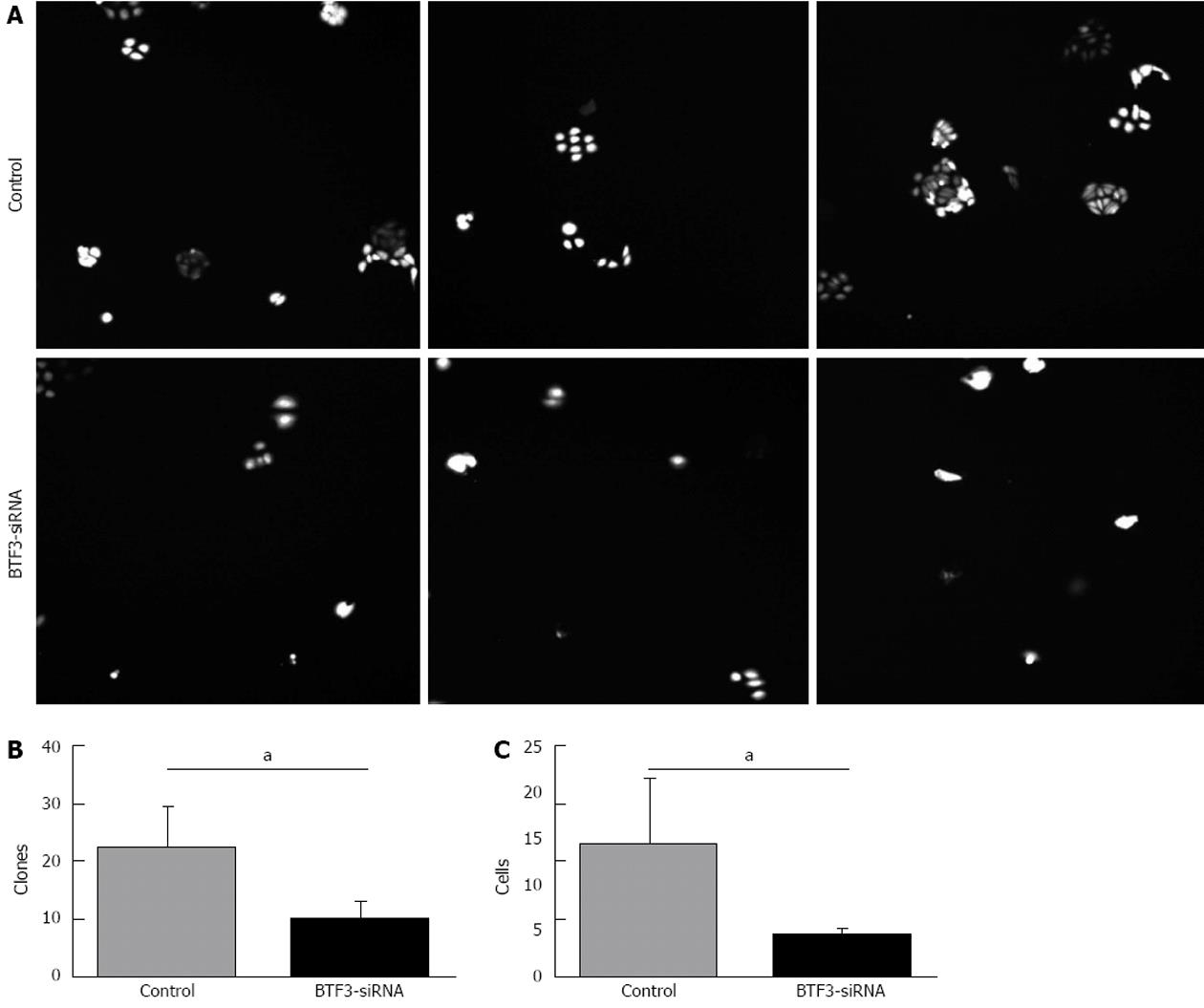
Figure 6 Colony forming abilities identified by Cellomic Assay System assays.
A: Colonies formed from control and basic transcription factor 3 (BTF3)-small interfering RNA (siRNA) transfected cells were imaged via the Cellomic Assay System; B: Number of clones plotted in a bar chart; C: Colony cell numbers of control and BTF3-siRNA transfected cells plotted in a bar chart. aP < 0.05 between groups were analysed with unpaired t test.
- Citation: Liu Q, Zhou JP, Li B, Huang ZC, Dong HY, Li GY, Zhou K, Nie SL. Basic transcription factor 3 is involved in gastric cancer development and progression. World J Gastroenterol 2013; 19(28): 4495-4503
- URL: https://www.wjgnet.com/1007-9327/full/v19/i28/4495.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i28.4495