Copyright
©2012 Baishideng Publishing Group Co.
World J Gastroenterol. Sep 7, 2012; 18(33): 4522-4532
Published online Sep 7, 2012. doi: 10.3748/wjg.v18.i33.4522
Published online Sep 7, 2012. doi: 10.3748/wjg.v18.i33.4522
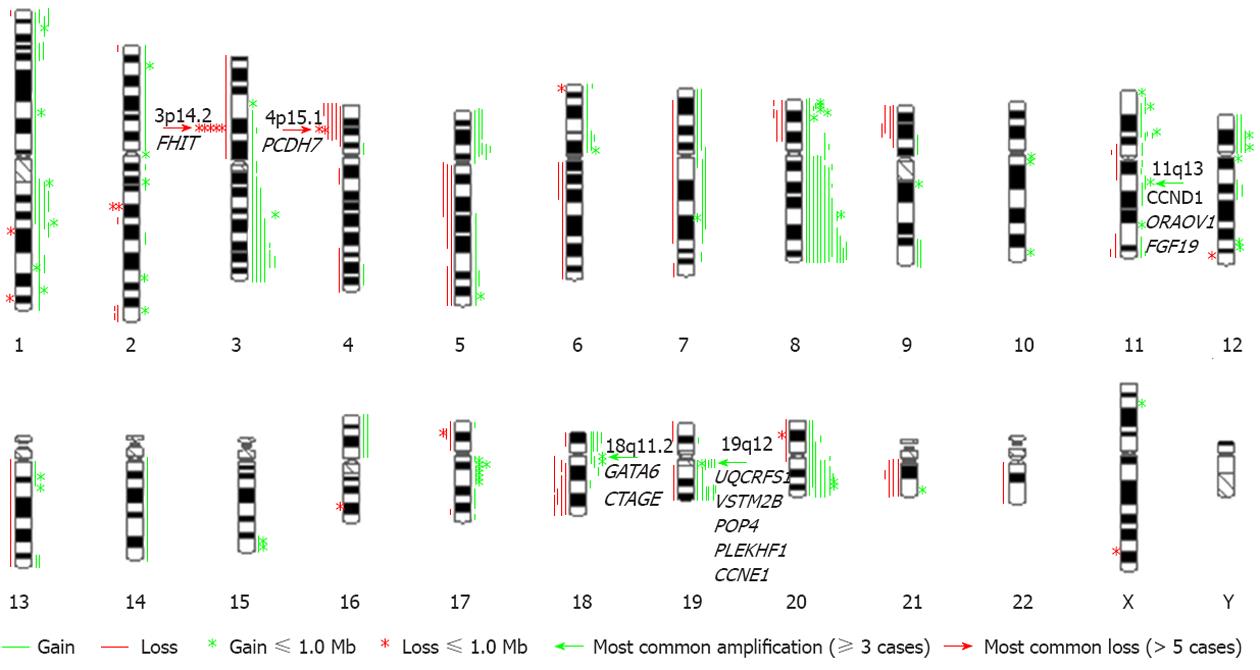
Figure 1 An overview of genomic imbalances in 20 primary gastric adenocarcinomas.
The total number of gains was 180 (54 ≤ 1 Mb), and the total number of losses was 70 (16 ≤ 1 Mb). Partial or whole gains of 8q (12/20), 19q (8/20), and 20q (8/20) were most frequent. The most common amplicons were of the 11q13, 18q11.2 and 19q12 regions (green arrows). The loss of the FHIT gene and the partial loss of 4p with the smallest region of overlap including the PCDH7 gene were the most frequent losses (red arrow).
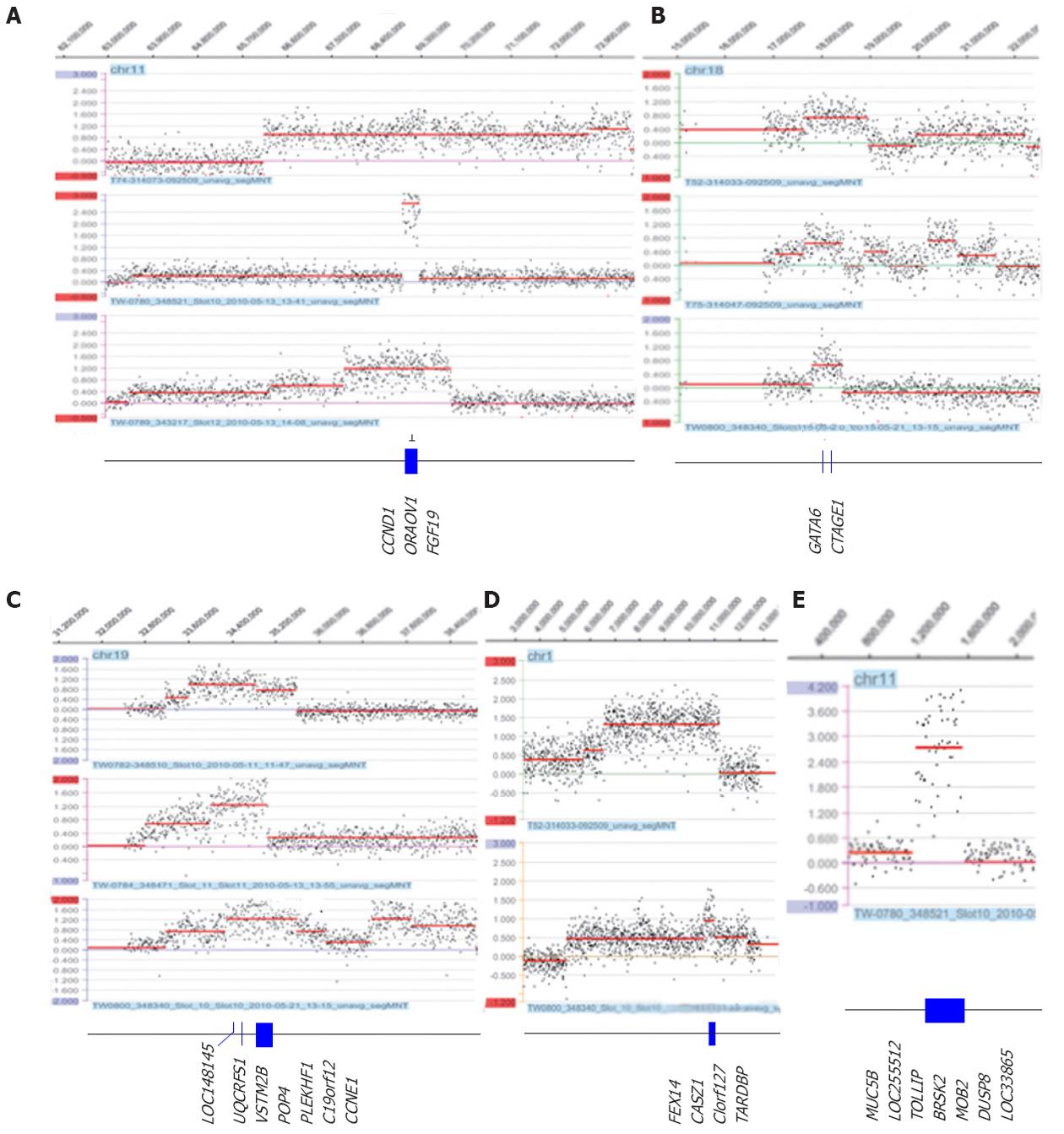
Figure 2 Representative amplifications detected by array comparative genomic hybridization and the genes that are located in the smallest region of overlap.
The most common amplicons at 11q13 (A), 18q11.2 (B) and 19q12 (C) and novel amplicons at 1p36.22 (D) and 11p15.5 (E) region (log2 > 0.5). The x-axis indicates the genomic location, and the y-axis indicates the log2 ratio.
Figure 3 Intergenic loss of FHIT/FRA3B on 3p14.
2 (A) and PDCH7 on 4p15.1 (B) detected by array comparative genomic hybridization. The x-axis indicates the genomic location, and the y-axis indicates the log2 ratio.
- Citation: Liu YY, Chen HY, Zhang ML, Tian D, Li S, Lee JY. Loss of fragile histidine triad and amplification of 1p36.22 and 11p15.5 in primary gastric adenocarcinomas. World J Gastroenterol 2012; 18(33): 4522-4532
- URL: https://www.wjgnet.com/1007-9327/full/v18/i33/4522.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i33.4522