Copyright
©2011 Baishideng Publishing Group Co.
World J Gastroenterol. Jul 14, 2011; 17(26): 3151-3157
Published online Jul 14, 2011. doi: 10.3748/wjg.v17.i26.3151
Published online Jul 14, 2011. doi: 10.3748/wjg.v17.i26.3151
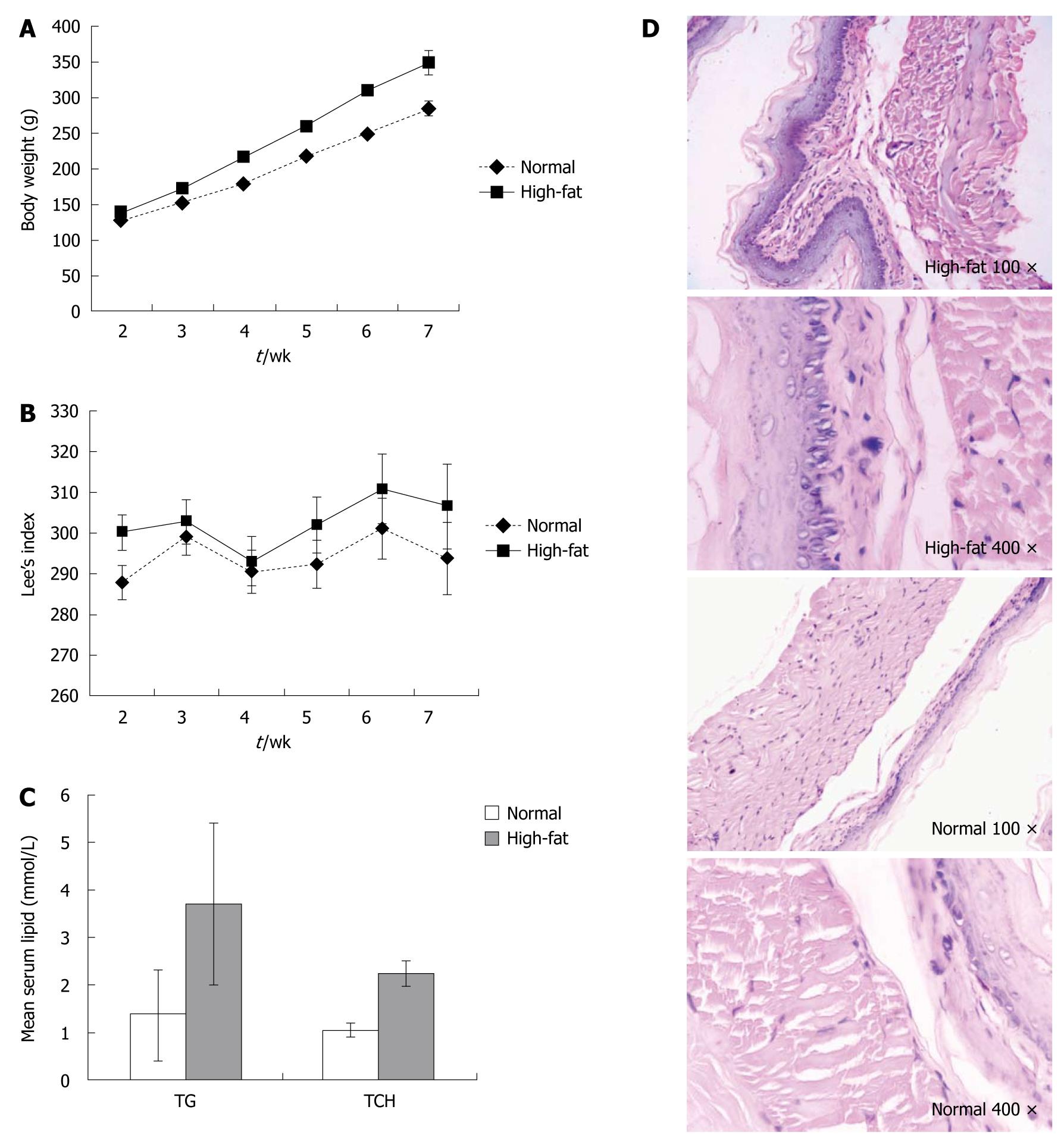
Figure 1 No significant change in distal esophageal mucosa in Sprague-Dawley rats fed a high-fat diet.
A: Body weight changes of age- and sex-matched rats in the high-fat diet and normal control groups (n = 10 each); B: Lees index changes for the rats in (A) (n = 10); C: Mean serum lipid for the rats in (A); D: Histological analysis of representative distal esophagus from the rats in (A) (Original magnification, hematoxylin-eosin staining, 100 × or 400 ×).
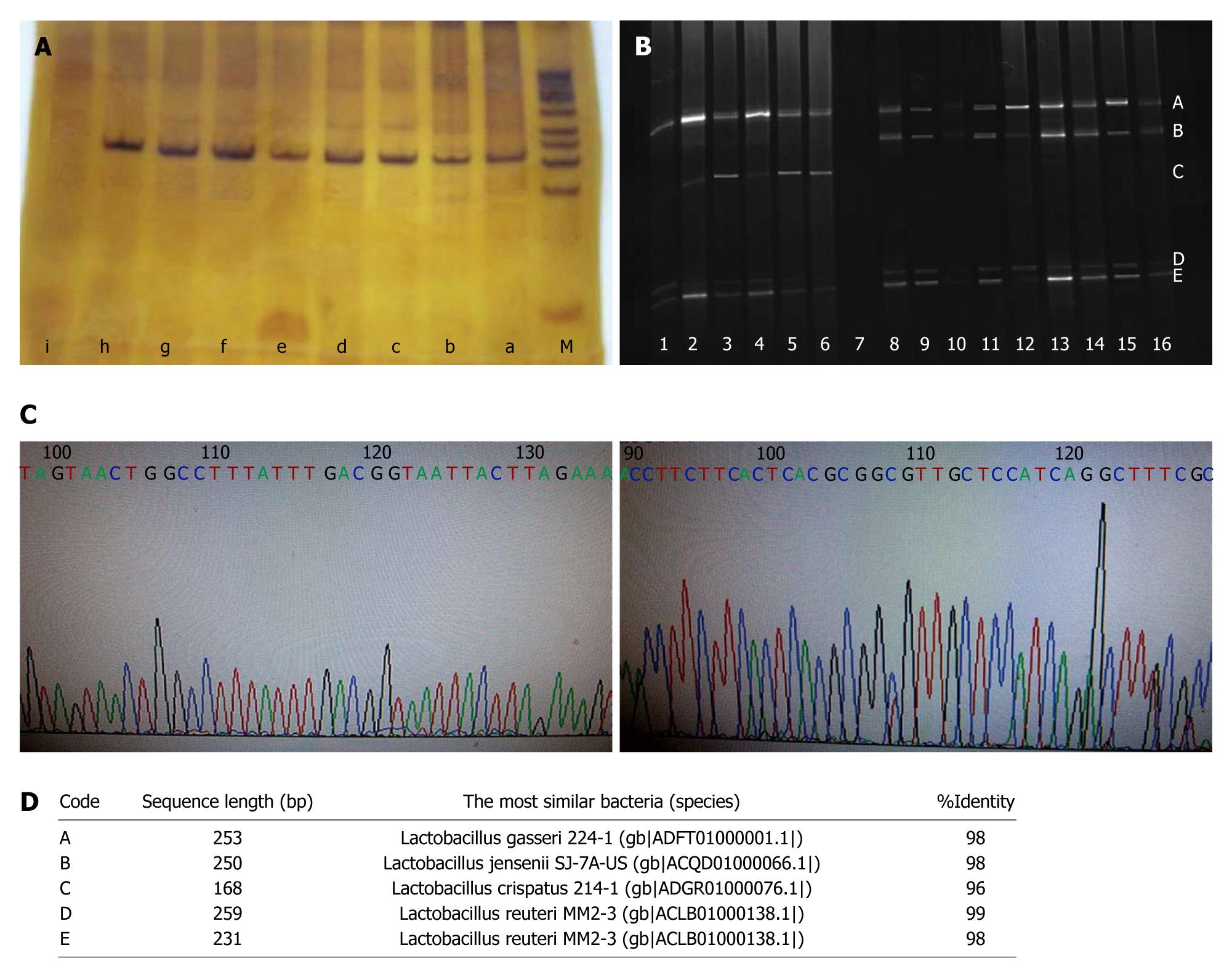
Figure 2 Determination of Lactobacillus species shift in distal esophagus of high-fat diet-fed Sprague-Dawley rats based on the combination of denaturing gradient gel electrophoresis and sequencing.
A: 16S rDNA of cultivable Lactobacillus was amplified by polymerase chain reaction using universal bacteria primers HAD1-GC and HAD2 in the high-fat diet and common diet groups. a: Lactobacillus crispatus (L. crispatus) for positive control, b-d: High-fat diet group; e-h: Common diet group; i: Negative control; M: DNA marker; B: Amplified products of 16S rDNA were separated by denaturing gradient gel electrophoresis (DGGE). Lanes 1-6: High-fat diet group; lane 7: Negative control; lanes 8-16: Normal diet group. The bands were marked with A, B, C, D and E, respectively; C: Purified bands were purified and sequenced; D: BLASTed online with V2-V3 region. Bands A, B, C, D and E represented Lactobacillus gasseri, Lactobacillus jensenii, L. crispatus, Lactobacillus reuteri, and Lactobacillus reuteri, respectively.
-
Citation: Zhao X, Liu XW, Xie N, Wang XH, Cui Y, Yang JW, Chen LL, Lu FG.
Lactobacillus species shift in distal esophagus of high-fat-diet-fed rats. World J Gastroenterol 2011; 17(26): 3151-3157 - URL: https://www.wjgnet.com/1007-9327/full/v17/i26/3151.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i26.3151