Copyright
copy;2010 Baishideng Publishing Group Co.
World J Gastroenterol. Sep 7, 2010; 16(33): 4135-4144
Published online Sep 7, 2010. doi: 10.3748/wjg.v16.i33.4135
Published online Sep 7, 2010. doi: 10.3748/wjg.v16.i33.4135
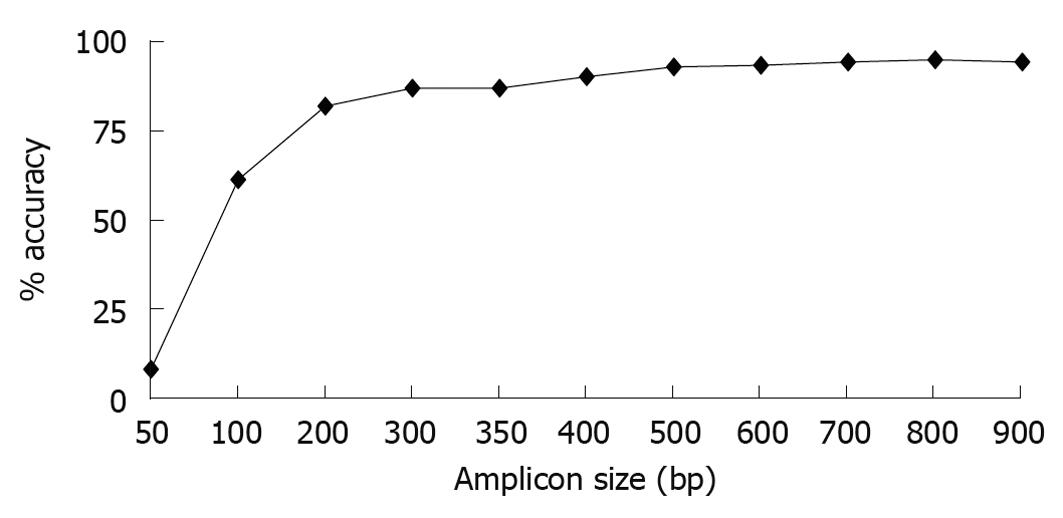
Figure 1 Classification accuracy is dependent on amplicon size.
Full length sequences were trimmed to 900, 800, 700, 600, 500, 400, 350, 300, 200,100, and 50 bases with each amplicon starting at Escherichia coli base 28. Each sequence trim was uploaded onto Ribosomal Database ProjectII classifier and the results at each taxonomical level were compared to results obtained using the full length sequence. Percent classification accuracies at the genus level for each amplicon size trim are shown.
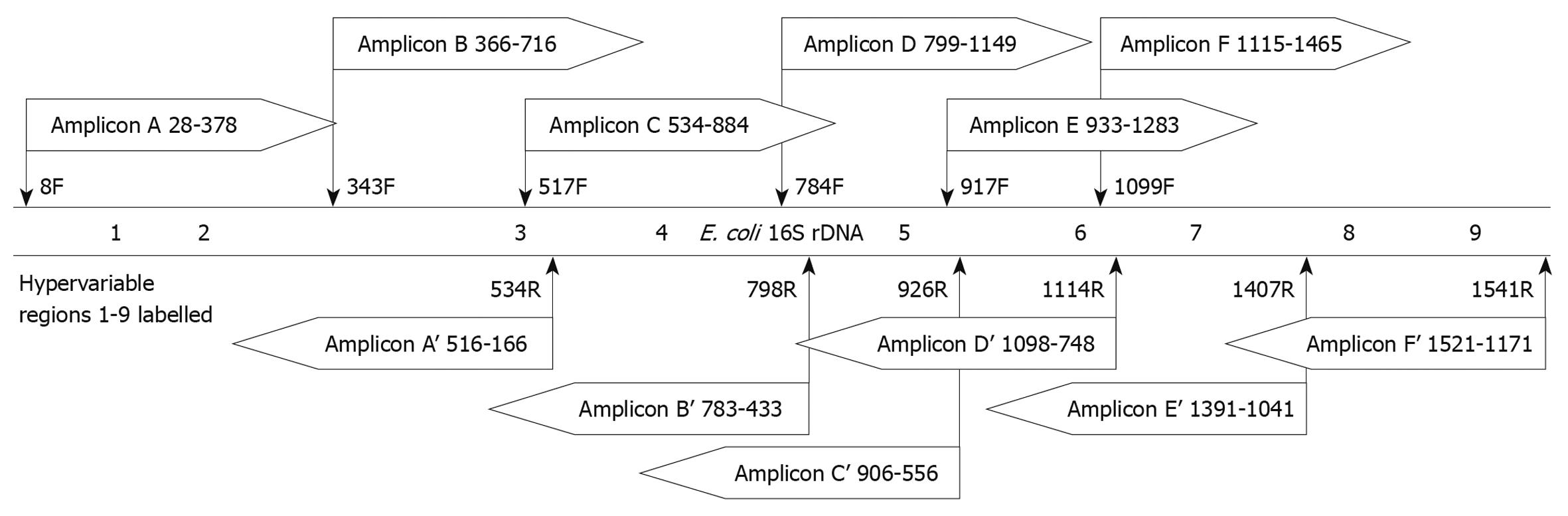
Figure 2 Design of amplicons for in silico evaluation.
Amplicons designed using the 6 universal primer sets as described in Table 3 were evaluated for theoretical forward and reverse reads. The schematic shows the relative position of each amplicon read and read direction, as well as which bases would be included in the sequence. The positional template is the Escherichia coli (E. coli) 16S rDNA gene with the approximate locations of the hypervariable regions labeled.
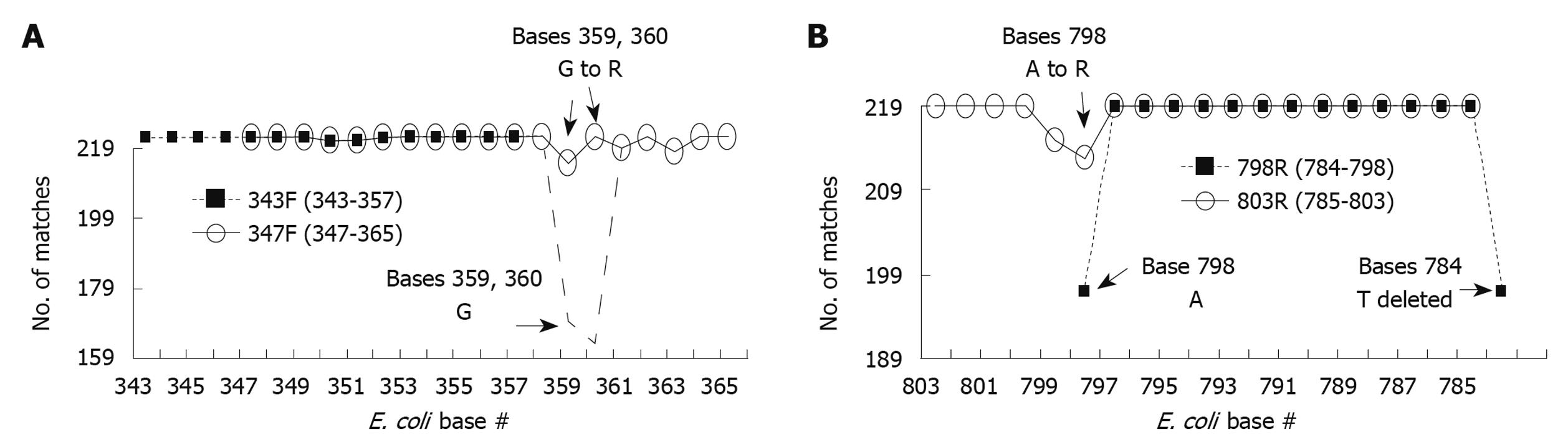
Figure 3 Optimization of primers used to generate amplicon B.
European Ribosome Database primers 343F (A) and 798R (B) were optimized to generate maximal % match with corresponding region in the 16S sequences of foregut species on a base by base manner. Each primer base was analyzed for the number of matches with the corresponding base of all 219 foregut species studied [base # assigned by position within Escherichia coli (E. coli) 16S sequence]. Most bases for both 343F and 798R showed 100% match (219/219), however some bases had slight mismatch % and a few bases had significant mismatch % (base 798, 784, and 359, 360). To have a better homology between the primers and the foregut 16S sequences, the bases with significant mismatch were adjusted to result in lower % mismatch. This was accomplished by changing 798A to R (increasing match from 197 to 213/219) and by changing 359G and 360G to R (increasing match from 169 and 163 to 212 and 219/219, respectively). Further modifications of primers were made to make them more suitable for polymerase chain reaction (PCR) reactions. R primer 5’ end was shifted from 798 to 803, and 3’ end from 784 to 785. F primer 5’ end was shifted from 343 to 347 and 3’ end from 357 to 365. These changes provided suitable melting and annealing temperatures for the designed primer pairs in our PCR reactions. Resulting primers were designated 347F and 803R.
- Citation: Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, DeSantis TZ, Brodie EL, Malamud D, Poles MA, Pei Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol 2010; 16(33): 4135-4144
- URL: https://www.wjgnet.com/1007-9327/full/v16/i33/4135.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i33.4135