Copyright
©2010 Baishideng.
World J Gastroenterol. Jan 14, 2010; 16(2): 201-209
Published online Jan 14, 2010. doi: 10.3748/wjg.v16.i2.201
Published online Jan 14, 2010. doi: 10.3748/wjg.v16.i2.201
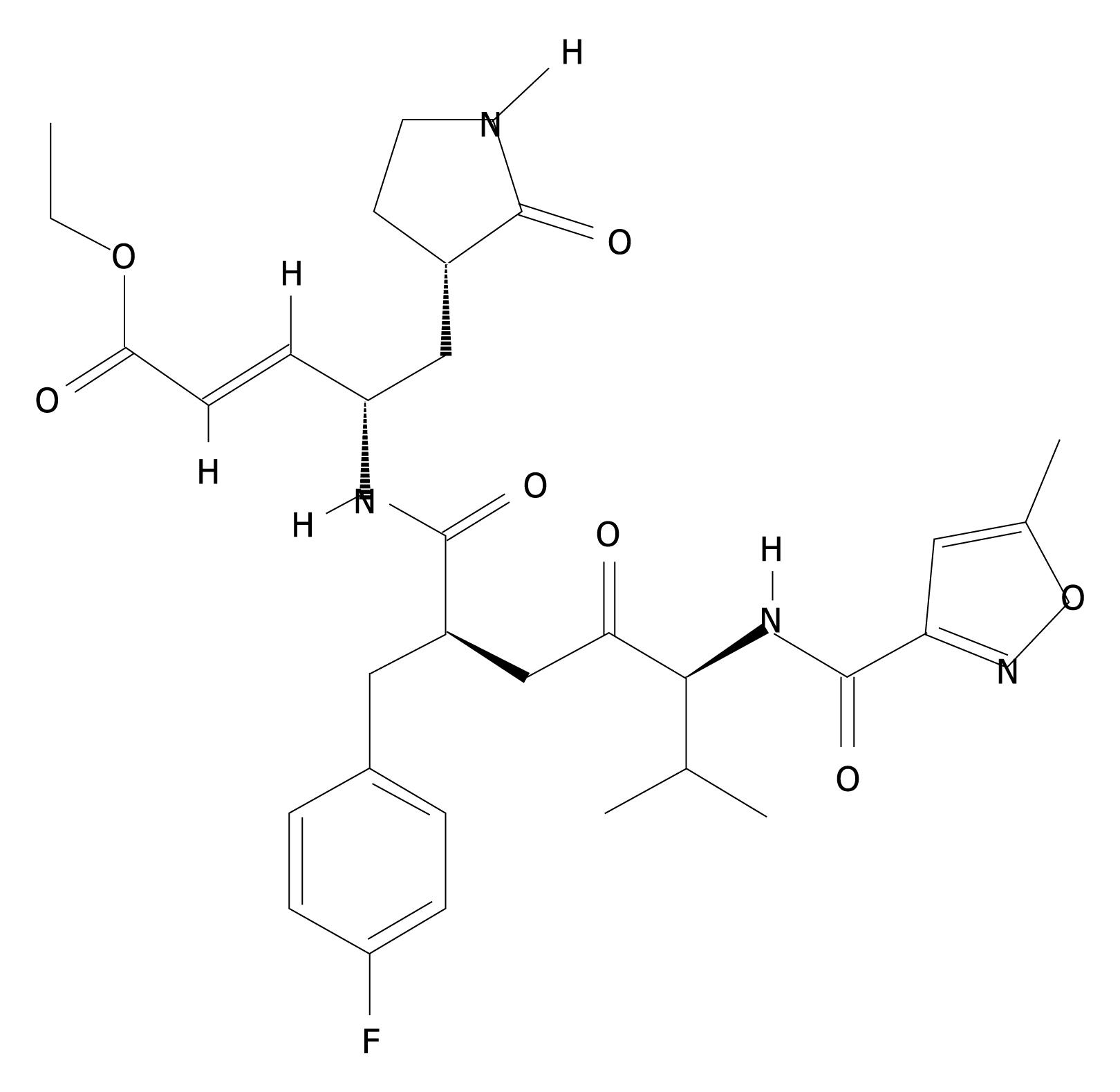
Figure 1 Chemical structure of rupintrivir.
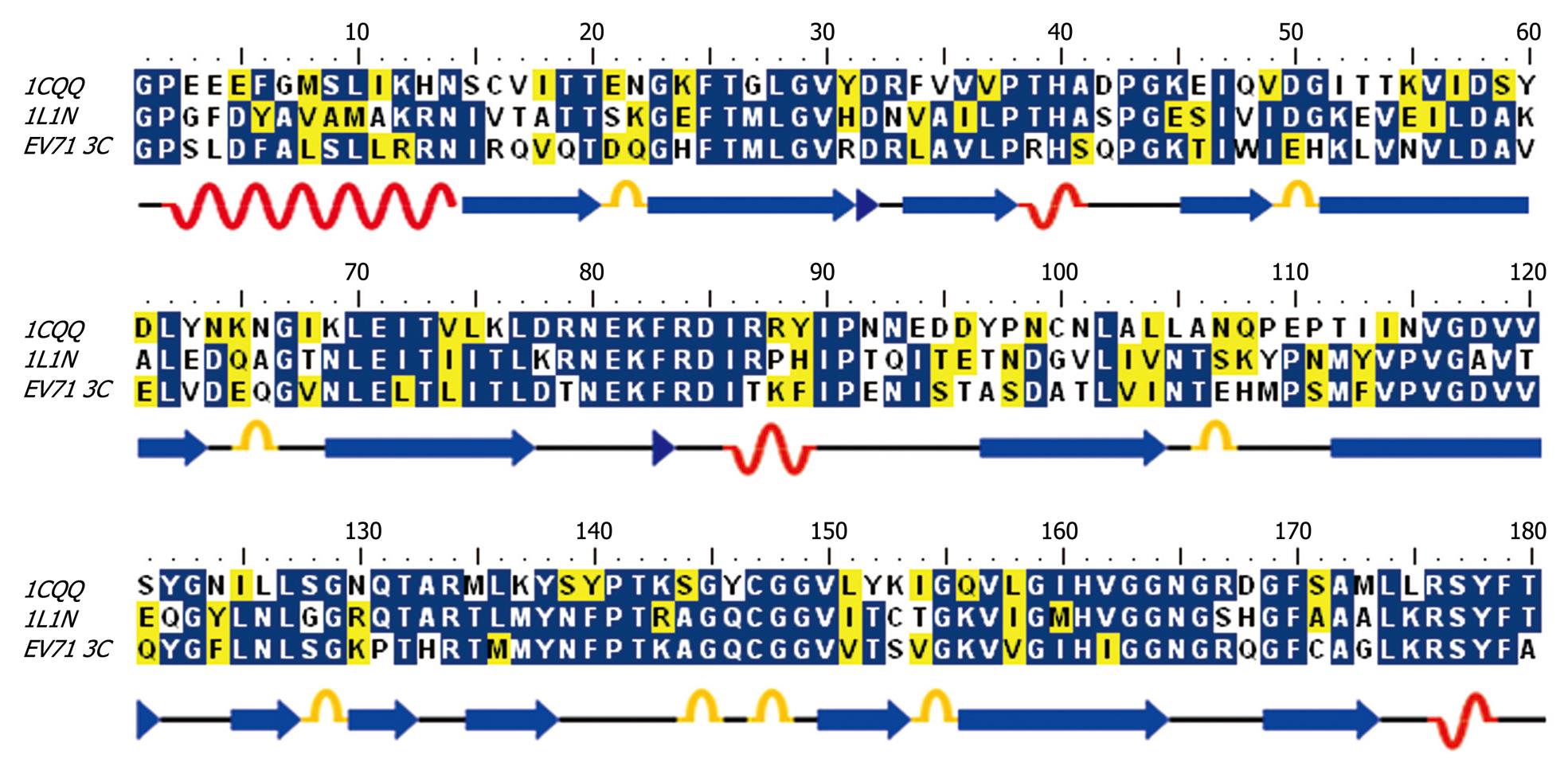
Figure 2 Sequence alignment of EV71 3C protease with HRV 3C (1CQQ) and Poliovirus 3C (1L1N).
Secondary structures (α helixes and β sheets) were illustrated.
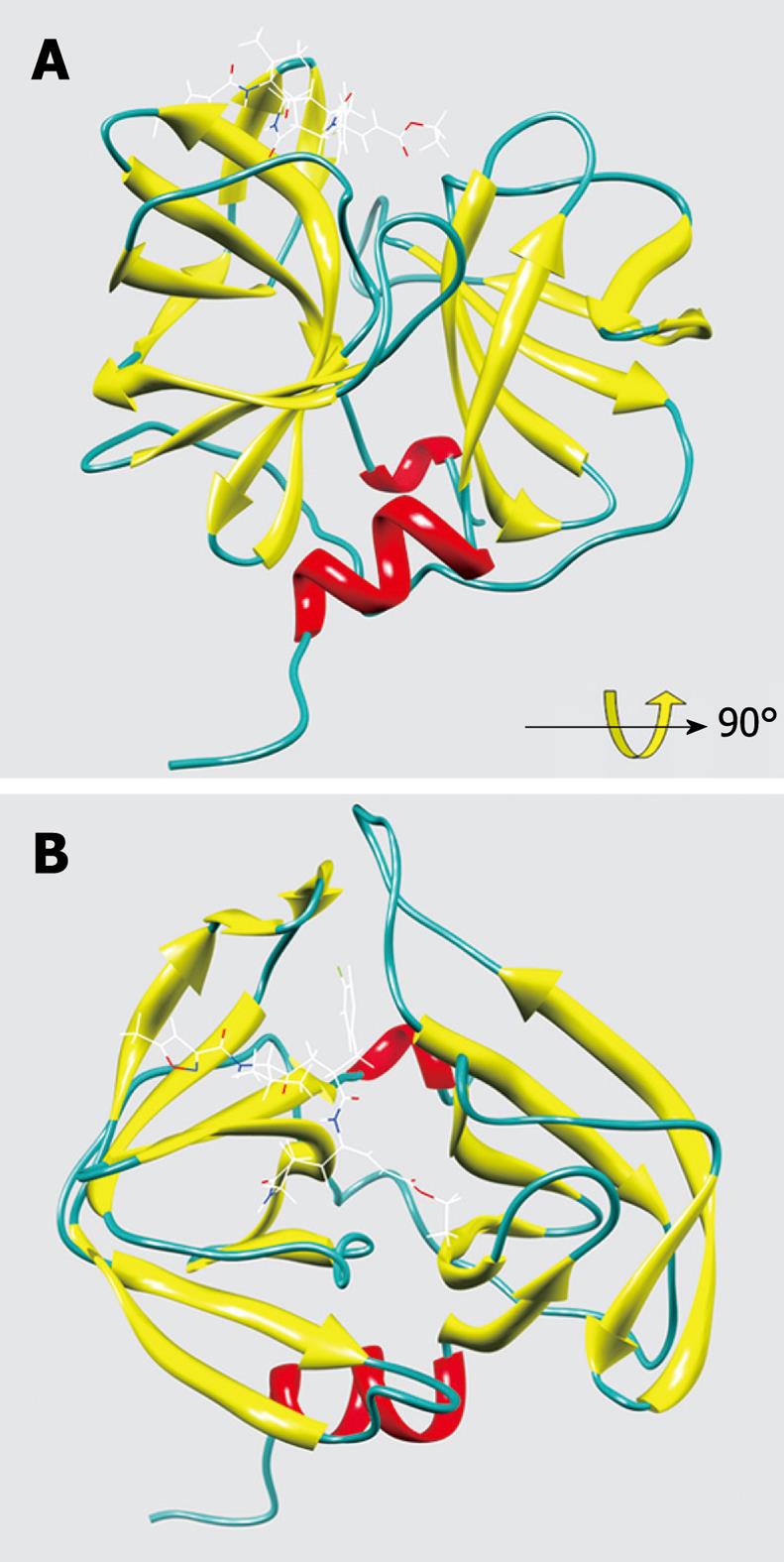
Figure 3 The overall structure of EV71 3C protease complexed with rupintrivir.
A: A solid ribbon presentation of EV71 3C with rupintrivir; B: 90° rotation view of the structure.
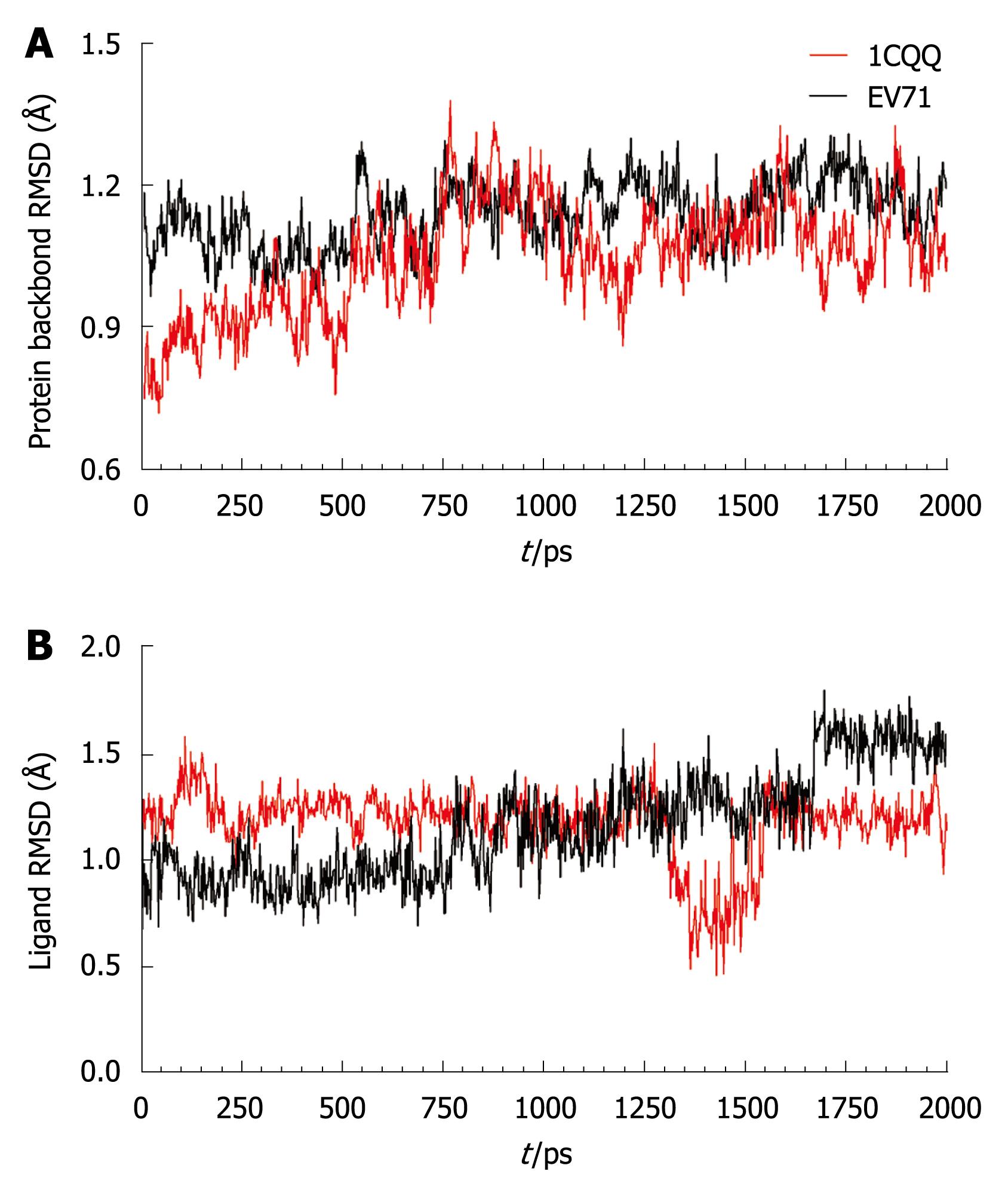
Figure 4 Receptor and ligand backbone displacement.
A: Backbone root mean square deviation (RMSD) of the HRV 3C (1CQQ) and EV71 3C protein; B: Ligand backbone RMSD during the 2ns molecular dynamics simulation.
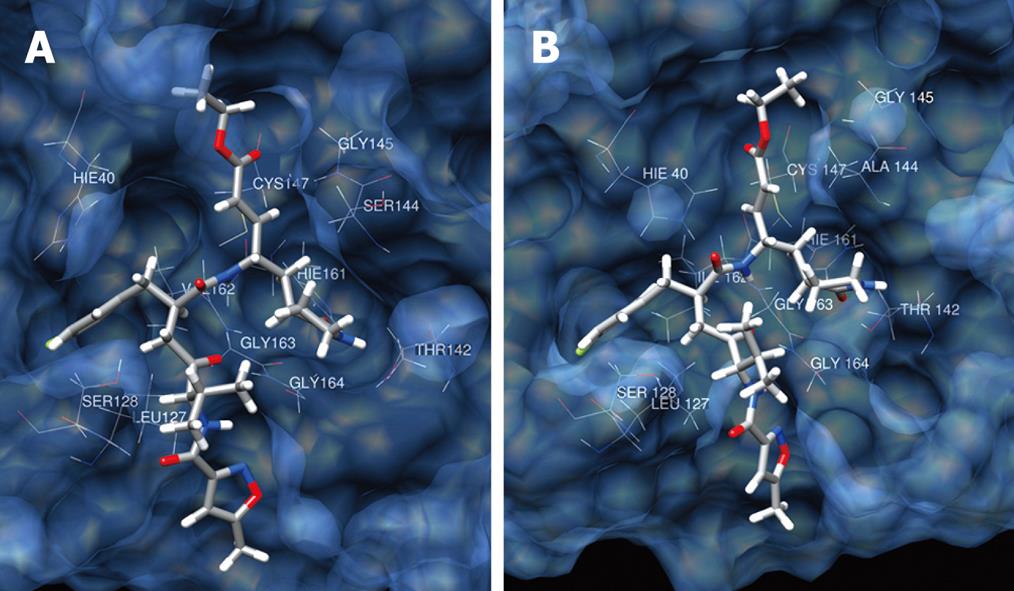
Figure 5 Binding mode of AG7088 with 1CQQ (A) and EV71 3C (B).
The protein surface is rendered semitransparent with associated backbone and side chain atom.
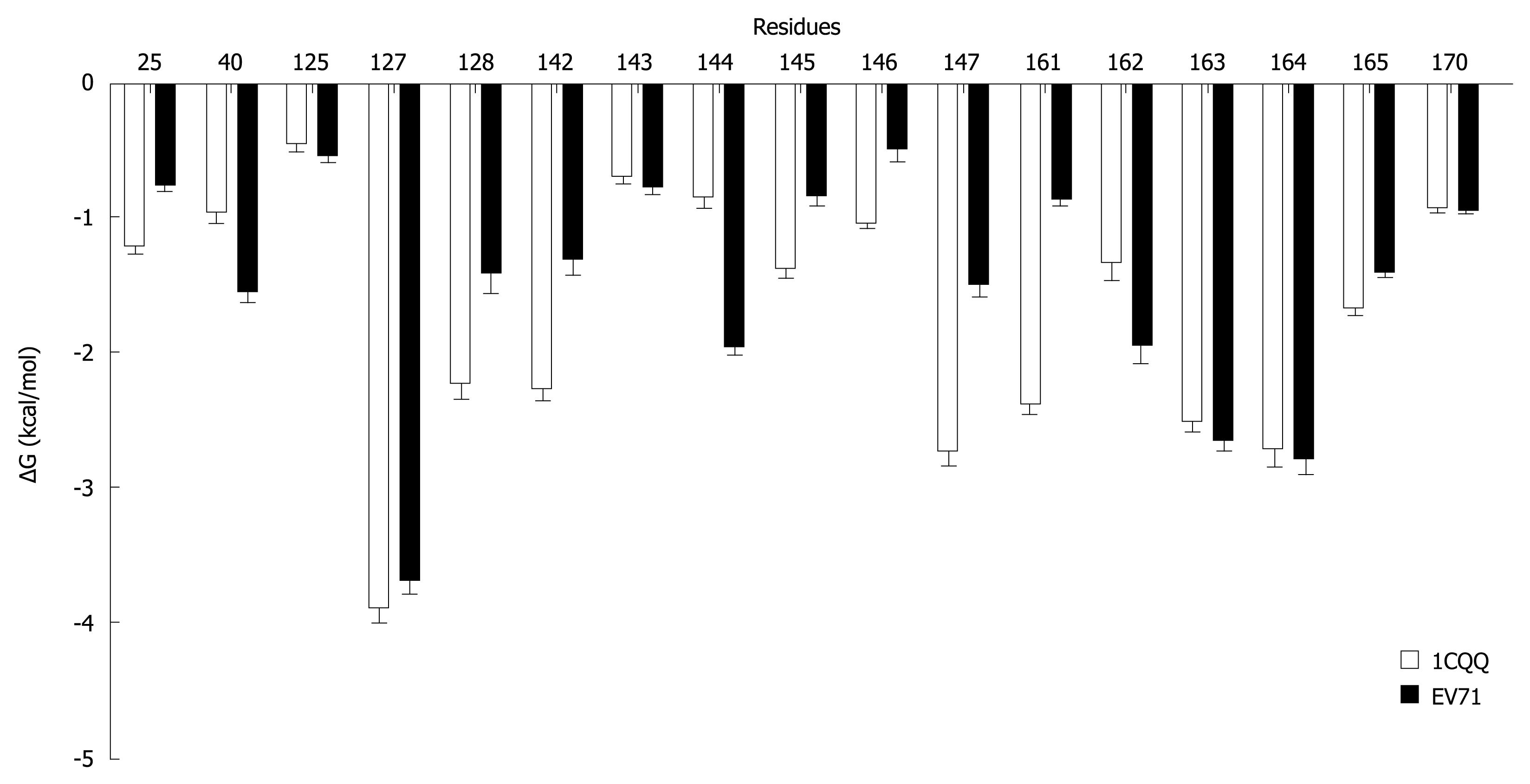
Figure 6 Energy decomposition (using MM-GBSA) of the key interaction residues in 1CQQ and EV71 3C complexes.
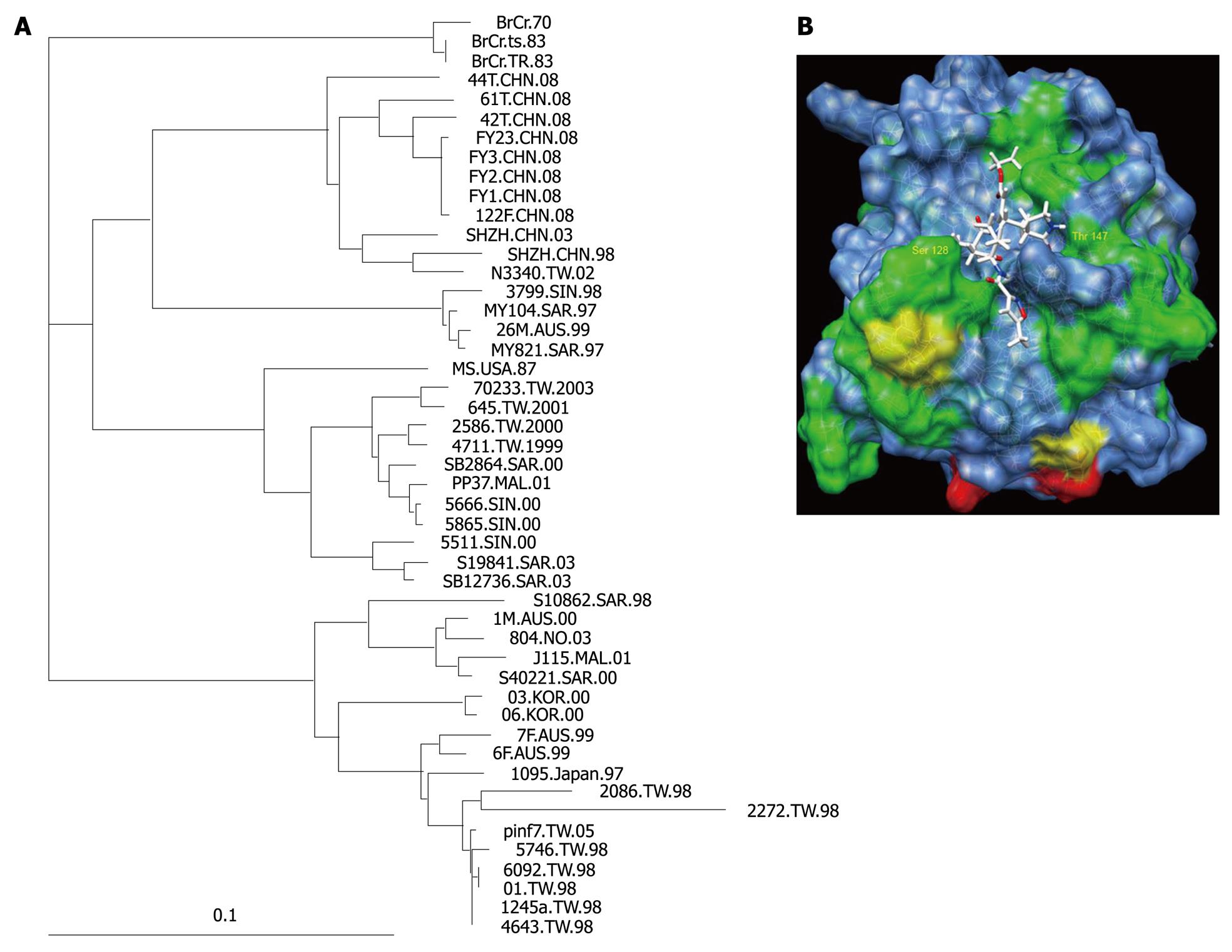
Figure 7 Sequence variability of EV71 3C protease and its effect on rupintrivir binding (A) Phylogenetic tree of 48 isolates of EV71 in various outbreaks.
(B) A colored representation of sequence variation on the surface of 3C protease. Positions with two, three and over three different residues are labeled green, yellow and red, respectively. Residue Ser 128 and Thr 142 were labeled.
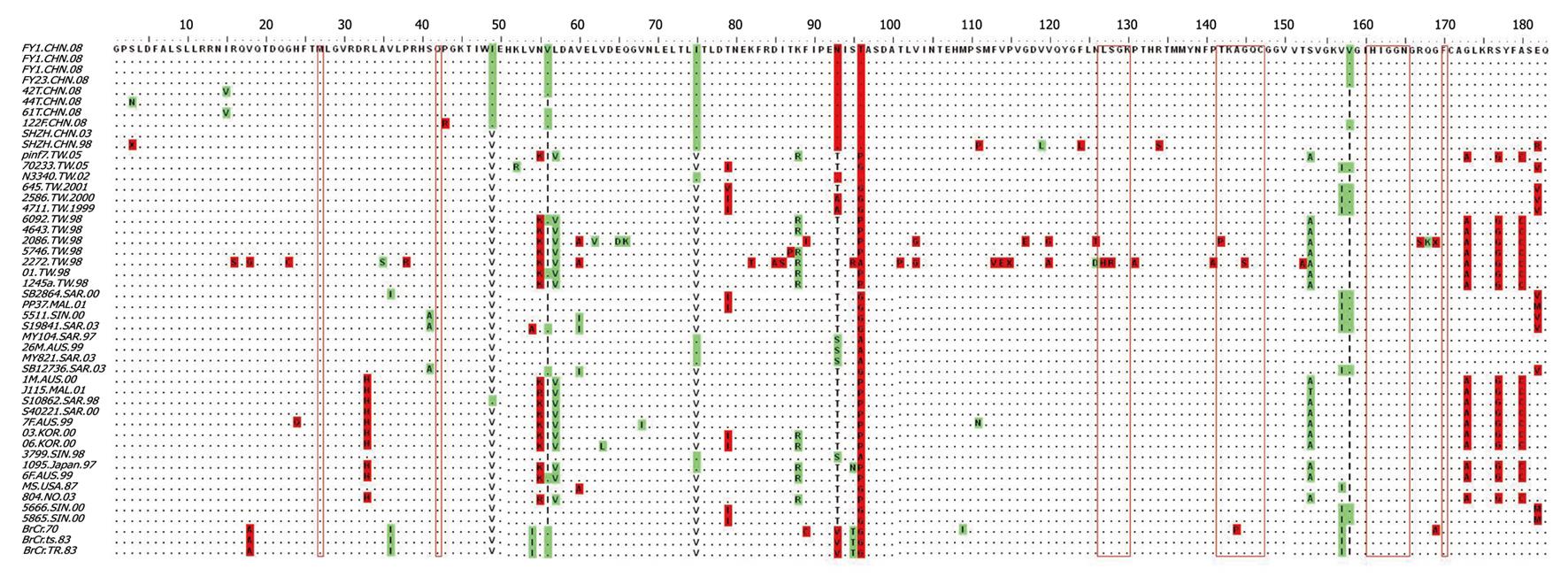
Figure 8 Sequence alignment of 3C protease from isolates collected in various epidemics (CHN: Mainland China; TW: Taiwan, China; FY: Fuyang, Anhui Province, China; SHZN: Shenzhen, China; AUS: Australia; SAR, SIN, MAL: Malaysia; KOR: Korea; NO: Norway).
Residues important for rupintrivir are labeled with red open boxes.
- Citation: Zhang XN, Song ZG, Jiang T, Shi BS, Hu YW, Yuan ZH. Rupintrivir is a promising candidate for treating severe cases of Enterovirus-71 infection. World J Gastroenterol 2010; 16(2): 201-209
- URL: https://www.wjgnet.com/1007-9327/full/v16/i2/201.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i2.201