Copyright
©2009 The WJG Press and Baishideng.
World J Gastroenterol. Aug 14, 2009; 15(30): 3799-3806
Published online Aug 14, 2009. doi: 10.3748/wjg.15.3799
Published online Aug 14, 2009. doi: 10.3748/wjg.15.3799
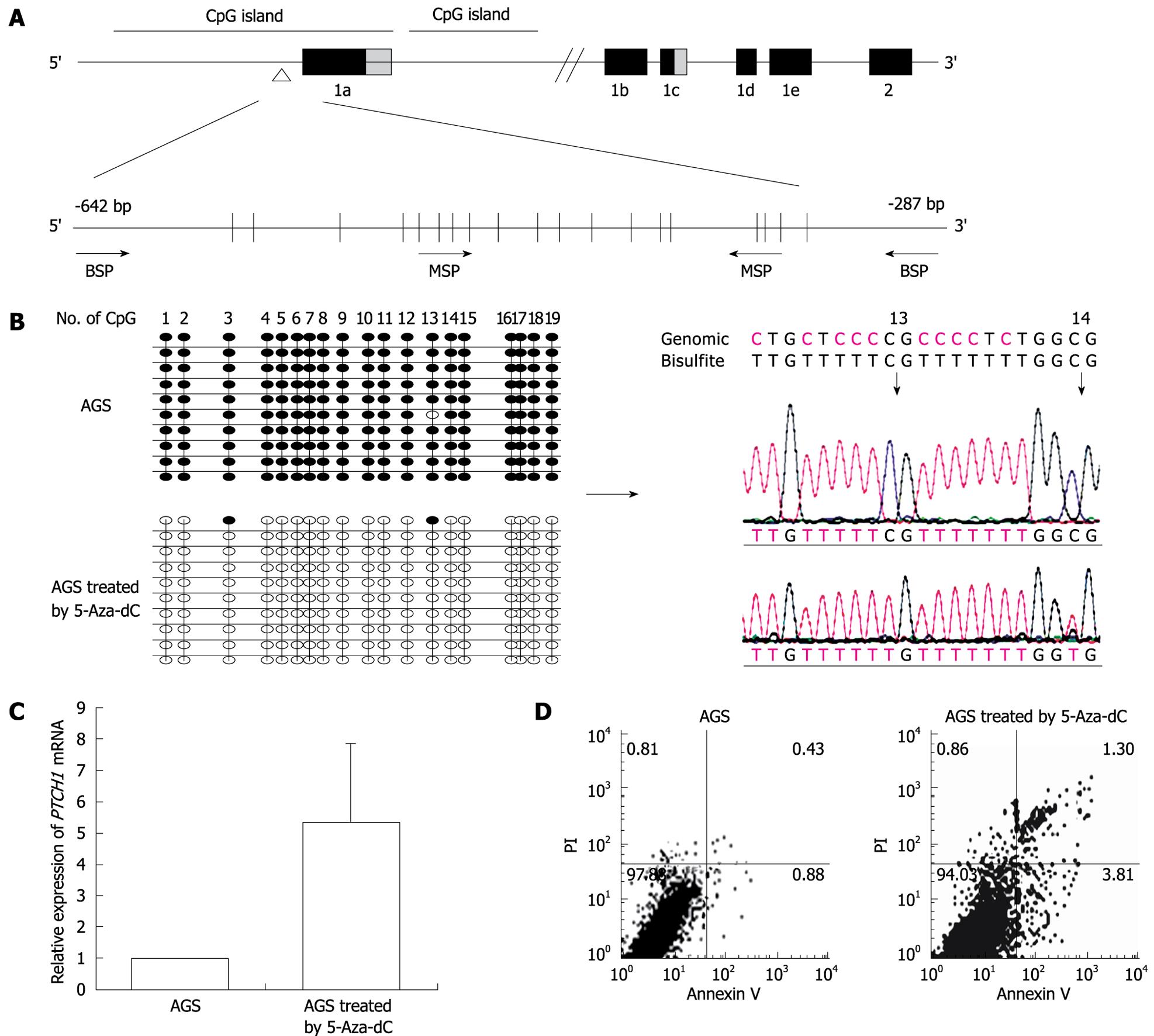
Figure 1 Analysis of methylation and expression of PTCH1a in gastric cancer cell line AGS.
A: Illustration of PTCH1a TRR and topology of MSP and BSP primers. BSP detection region contained 19 CpG sites, and MSP up-primer and down-primer contained four and three CpG sites, respectively. The detection amplicon is indicated by the empty triangle; B: Alteration of PTCH1a TRR methylation in gastric cancer cell line AGS treated with 5-Aza-dC. Genomic DNA from untreated AGS cells and those treated with 5-Aza-dC (1 μmol/L) were analyzed by BSP at day 4. The left column indicates alterations of the 19 CpG sites contained in the BSP amplicon through 10 cloned sequences after 5-Aza-dC treatment. The right column displays part of the sequence of the methylated and unmethylated clones. Black dot, methylated; white dot, unmethylated; C: Alteration of PTCH1 expression in gastric cancer cell line AGS treated with 5-Aza-dC. PTCH1 gene expression in AGS cells treated with 5-Aza-dC (1 μmol/L) at day 4 was detected by real-time PCR relative to untreated AGS cells. Expression of PTCH1 gene was enhanced significantly in the treated AGS cells compared with untreated (P < 0.01, n = 3, independent tests). Box, mean; bar, SD; D: Analysis of apoptosis of AGS cells treated with 5-Aza-dC. The rate of apoptosis in AGS cells treated with 5-Aza-dC (1 μmol/L) at day 4 was significantly higher compared to untreated cells (P < 0.01, n = 3). The representative analysis of AGS cell apoptosis by annexin V/PI method is shown.
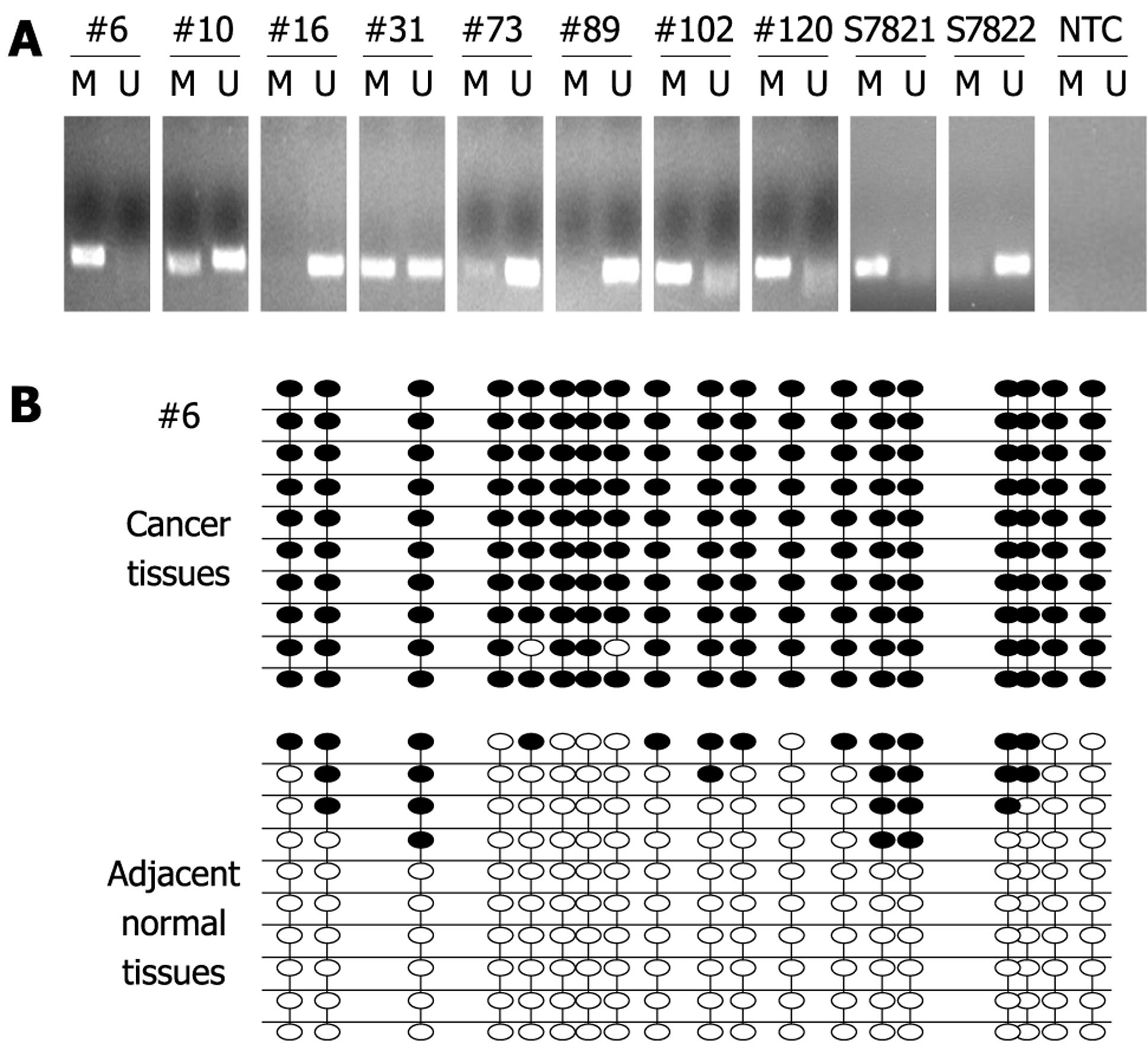
Figure 2 Methylation of PTCH1a TRR in gastric cancer tissues.
A: Methylation of PTCH1a TRR in gastric cancer tissues (n = 170) using MSP. MSP results from eight representative patients (#) are shown. The DNA bands in lanes labeled with M represent the products amplified with the methylation-specific primers, while DNA bands labeled with U represent the products amplified with the non-methylation-specific primers. CpGenome Universal Methylated DNA (S7821) and the CpGenome Universal Unmethylated DNA (s7822) were used as controls for methylation and non-methylation. Water was used as non-template control (NTC); B: Genomic DNA of gastric cancer tissues and its corresponding normal tissues from a representative patient (#6) was analyzed by BSP. Methylation patterns of the 19 CpG sites contained in the BSP amplicon through 10 clone sequence analyses in cancer tissues, and corresponding normal tissues are shown. Black dot, methylation; white dot, non-methylation.
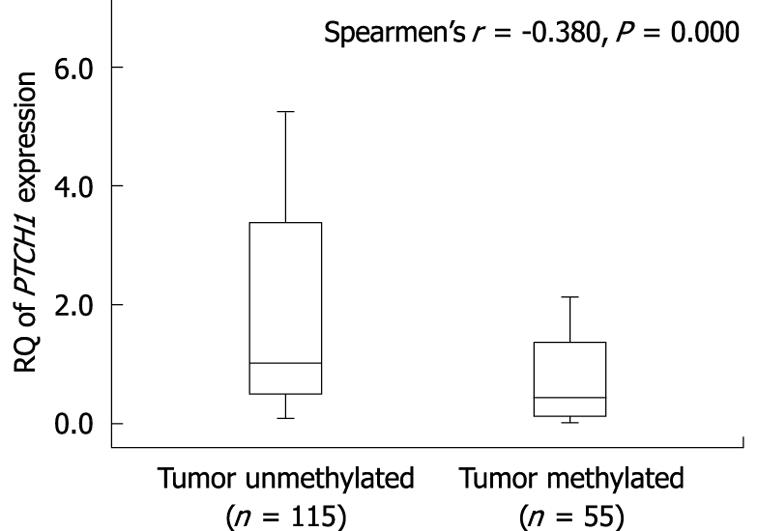
Figure 3 Correlation between methylation of PTCH1a gene TRR and expression of PTCH1 in gastric cancer tissues.
Box plot illustrating the loss of PTCH1 gene expression in relation to the methylation of PTCH1a gene TRR in human gastric cancer tissues (n = 170). The Y axis indicates the RQ value of PTCH1 gene mRNA expression was calculated in comparison with a calibrator (the expression level of pooled adjacent normal tissue samples). Horizontal lines: Group medians; Boxes: 25%-75% quartiles; Vertical lines: Range, peak and minimum.
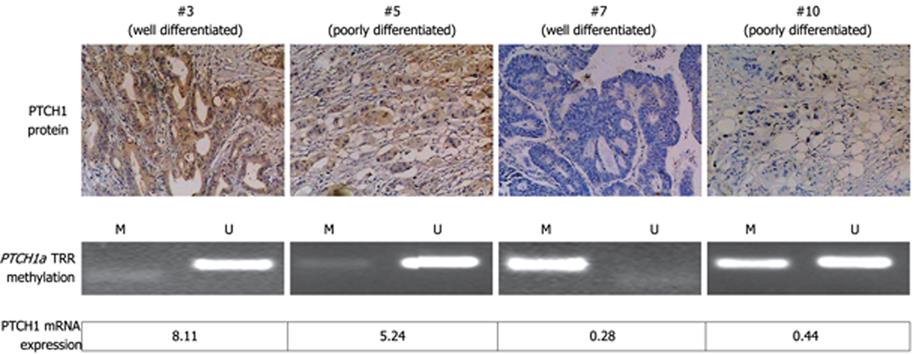
Figure 4 Correlation of PTCH1 mRNA and protein expression with methylation of PTCH1a TRR.
Expression of PTCH1 genes, as well as methylation of PTCH1a TRR are displayed in four representative gastric cancer tissue samples (#3, #5, #7 and #10). PTCH1 protein expression was detected by immunohistochemical staining (original magnification, × 100). Methylation of PTCH1a TRR was detected by MSP. The RQ value of PTCH1 gene mRNA expression was calculated in comparison with a calibrator (the expression level of pooled adjacent normal tissue samples). The well-differentiated tissue sample #3 and the poorly differentiated tissue sample #5 showed non-methylated products by MSP, positive expression of PTCH1 protein, and higher RQ value of PTCH1 mRNA expression. The well-differentiated tissue sample #7 and poorly differentiated tissue sample #10 showed methylated products by MSP, negative expression of PTCH1 protein, and lower RQ value of PTCH1 mRNA expression.
- Citation: Du P, Ye HR, Gao J, Chen W, Wang ZC, Jiang HH, Xu J, Zhang JW, Zhang JC, Cui L. Methylation of PTCH1a gene in a subset of gastric cancers. World J Gastroenterol 2009; 15(30): 3799-3806
- URL: https://www.wjgnet.com/1007-9327/full/v15/i30/3799.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.3799