Copyright
©2008 The WJG Press and Baishideng.
World J Gastroenterol. Sep 21, 2008; 14(35): 5419-5427
Published online Sep 21, 2008. doi: 10.3748/wjg.14.5419
Published online Sep 21, 2008. doi: 10.3748/wjg.14.5419
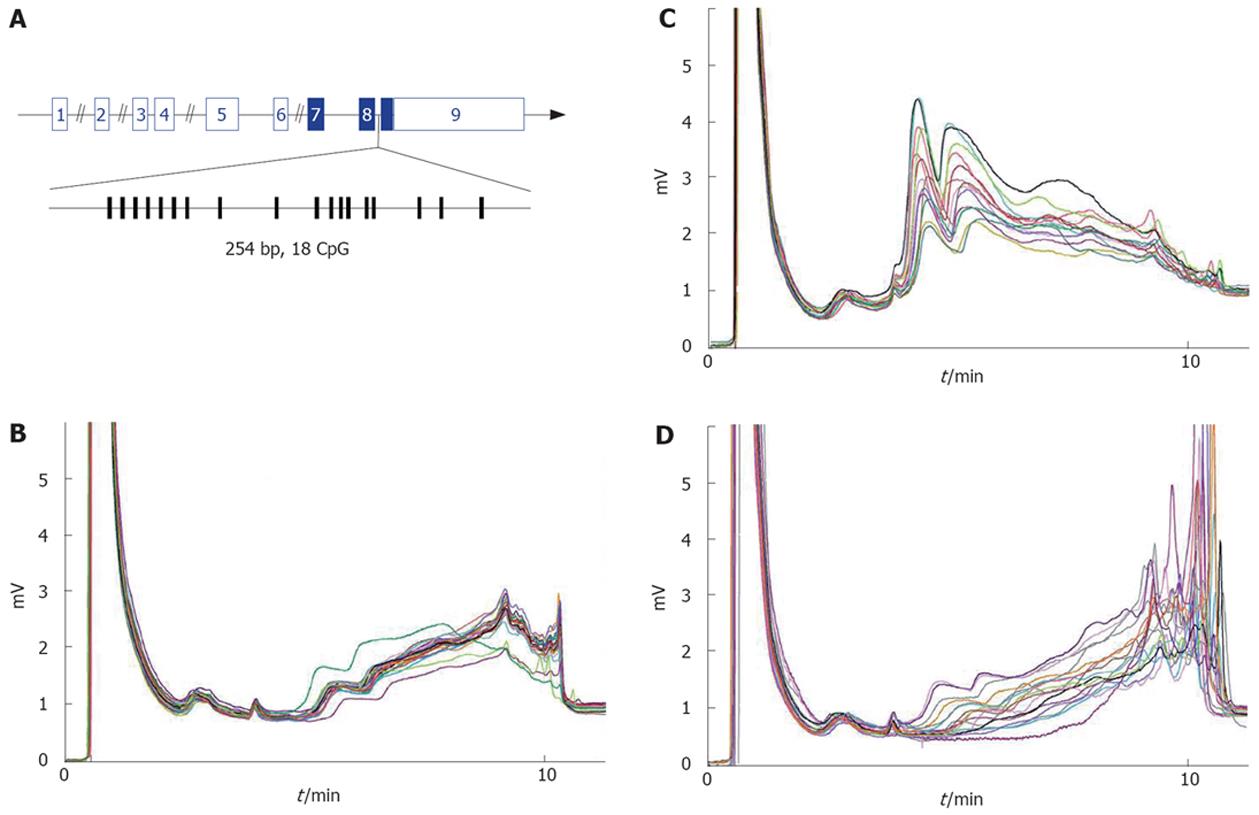
Figure 1 Methylation analysis of Igf2 DMR2 in normal and HCV related cirrhosis livers.
A: Exon-intron structure of Igf2 gene. Exons are shown as numbered boxes (plain are coding). The 254bp fragment of Igf2 DMR2 amplified for methylation analysis is enlarged below. Vertical lines indicate CpG positions. B: DHLPLC chromatograms of PCR products from normal liver samples. Twenty-two out of 25 are superimposable, and this major profile was used to assess hypermethylated profiles (ie more methylated than normal liver). C and D: DHPLC chromatograms of PCR products from HCV-related cirrhosis. Among 94 samples, 13 (C) and 17 (D) samples show respectively hypomethylated and hypermethylated profiles.
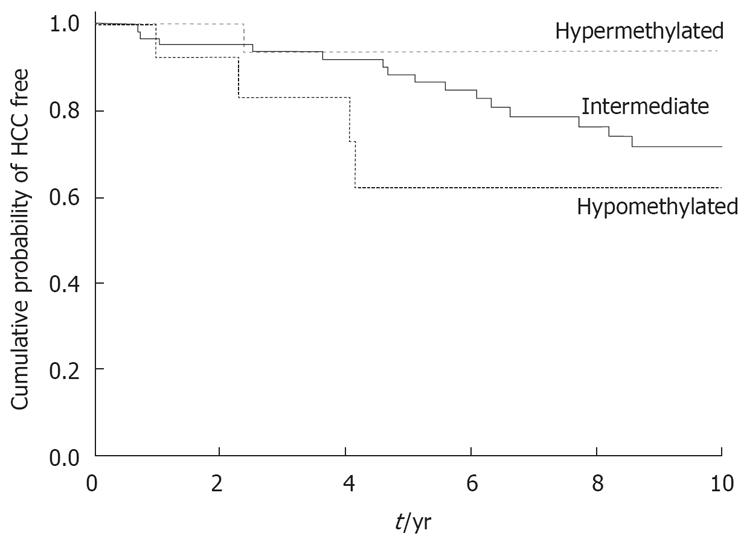
Figure 2 Occurrence of hepatocellular carcinoma at 10 years according to the methylation profile at the Igf2 gene in 94 patients with Child-Pugh A hepatitis C-related cirrhosis (Kaplan-Meier method).
Test of heterogeneity of HCC distributions: (log-rank test) P = 0.13. Test of the linear trend between levels of Igf2 methylation and corresponding survival functions: Breslow (Generalized Wilcoxon) P = 0.037.
-
Citation: Couvert P, Carrié A, Pariès J, Vaysse J, Miroglio A, Kerjean A, Nahon P, Chelly J, Trinchet JC, Beaugrand M, Ganne-Carrié N. Liver
insulin-like growth factor 2 methylation in hepatitis C virus cirrhosis and further occurrence of hepatocellular carcinoma. World J Gastroenterol 2008; 14(35): 5419-5427 - URL: https://www.wjgnet.com/1007-9327/full/v14/i35/5419.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.5419