Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 28, 2007; 13(4): 497-502
Published online Jan 28, 2007. doi: 10.3748/wjg.v13.i4.497
Published online Jan 28, 2007. doi: 10.3748/wjg.v13.i4.497
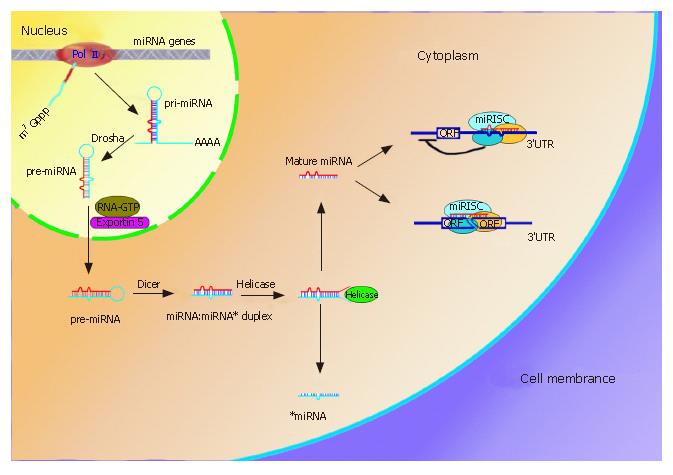
Figure 1 miRNAs biogenesis and two mechanisms involved in gene expression.
MiRNAs genes often cluster on the chromosome and are transcribed by RNA PolII to form pri-miRNAs in the nucleus. The pri-miRNAs then undergo procession by RNase III, Drasha, and are exported to cytoplasm by Exportin 5. Another RNase III, Dicer, further processes the pre-miRNA to generate a ~22nt miRNA:miRNA* duplex, where miRNA* is complementary to miRNA. Helicase can divide the duplex into two separate ones. Whereas miRNA* is degraded, mature miRNA can enter the the miRNA-induced silence complex (miRISC). The miRISC complex block protein synthesis by imperfectly binding to the 3’UTR of the mRNA (upper right), the other one is to endonucleolytically cleave the target mRNA by perfect or nearly perfect base pairing (lower right).
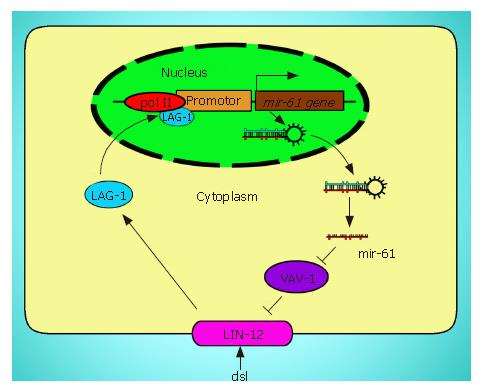
Figure 2 The positive loop between mir-16 and LIN-12.
Exogenous signal activates LIN-12/Notch signaling pathway, which promotes the transcription of mir-61. Overexpression of mir-61 inhibits the activity of VAV-1 (purple) that reduces the activity of LIN-12 (pink), where LIN-12, mir-61, and VAV-1 form a feedback loop that helps maximize LIN-12 activity.
- Citation: Liu W, Mao SY, Zhu WY. Impact of tiny miRNAs on cancers. World J Gastroenterol 2007; 13(4): 497-502
- URL: https://www.wjgnet.com/1007-9327/full/v13/i4/497.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i4.497