Copyright
©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Jul 7, 2006; 12(25): 4009-4013
Published online Jul 7, 2006. doi: 10.3748/wjg.v12.i25.4009
Published online Jul 7, 2006. doi: 10.3748/wjg.v12.i25.4009
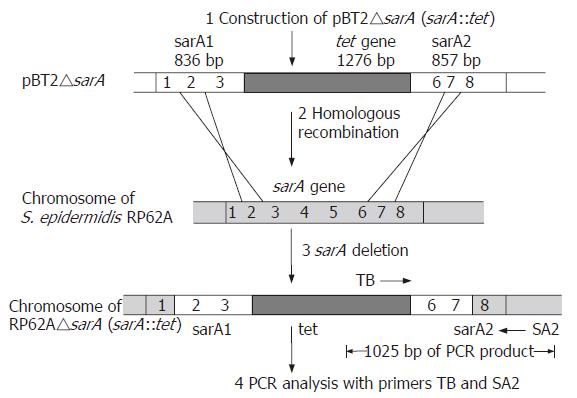
Figure 1 Disruption and detection of chromosomal sarA locus in S.
epidermidis. 1. Insertion of tet gene into sarA locus to form recombinant plasmid pBT△sarA. 2. Double-exchange homologous recombination between chromosomal DNA and plasmid DNA. 3. Locus sarA of chromosome was destroyed by DNA recombination. 4. Detection of recombination by PCR amplification with primers TB and SA2.
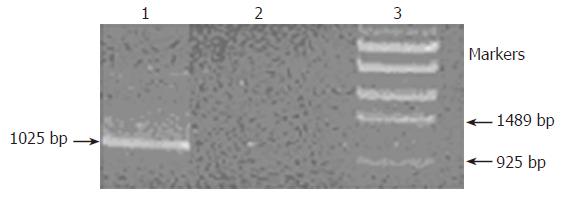
Figure 2 Detection of sarA gene locus in chromosome DNA by PCR with primers SA2 and TB.
Lane 1, The product was amplified from S. epidermidis RP62A△sarA with tet insertion; lane 2, No product was amplified from S. epidermidis RP62A without tet insertion; lane 3, DNA markers.
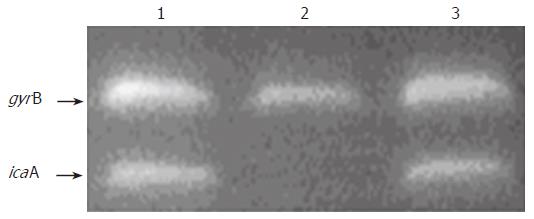
Figure 3 Identification of the genes icaA and gyrB transcription in S.
epidermidis by RT-PCR analysis. 1. S. epidermidis RP62A; 2. S. epidermidis RP62A△sarA;
Figure 4 Biofilm formation on the glass surface in the tube culturing S.
epidermidis.
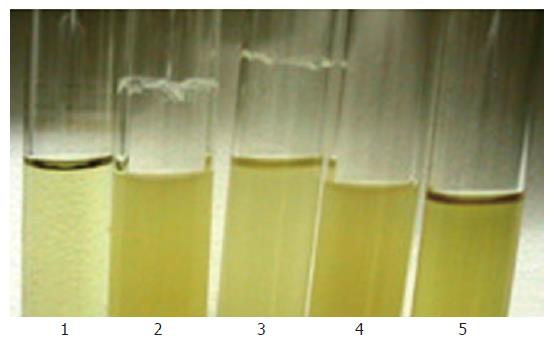
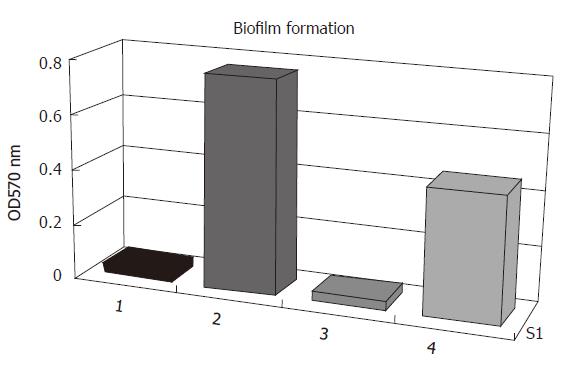
Figure 5 Biofilm formation on polystyrene tissue culture plates of S.
epidermidis in TSB medium. 1. biofilm-negative strain S. epidermidis ATCC12228; 2. original strain S. epidermidis RP62A; 3. sarA::tet insertion mutant S. epidermidis RP62A△sarA; 4. complemented strain S. epidermidis RP62A △sarA (pHPS9sarA).
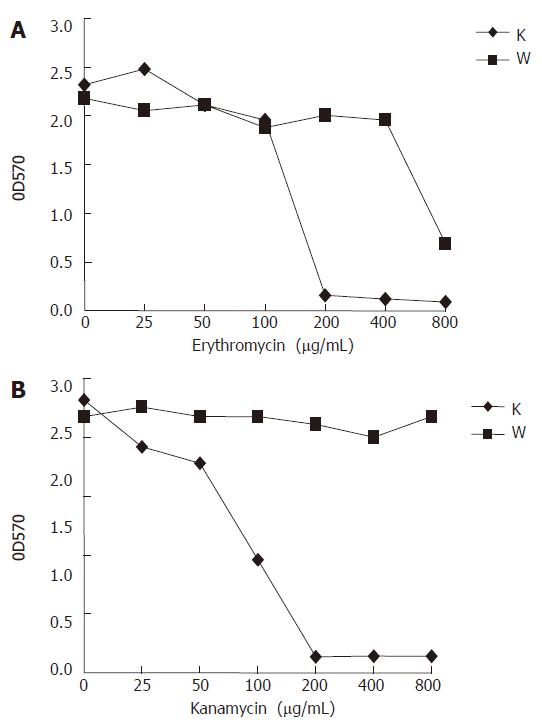
Figure 6 Response of RP62A (original) and RP62A△sarA (mutant) to two antibiotics in shaking tube cultures.
The cultures of the wild type RP62A (W) and the mutant RP62A△sarA (K) were incubated in a shaking incubator (250 r/min) at 37°C for 24 h, and diluted in fresh TSB medium. Determination of optical density (OD) values was carried out at 570 nm.
-
Citation: Tao JH, Fan CS, Gao SE, Wang HJ, Liang GX, Zhang Q. Depression of biofilm formation and antibiotic resistance by
sarA disruption inStaphylococcus epidermidis . World J Gastroenterol 2006; 12(25): 4009-4013 - URL: https://www.wjgnet.com/1007-9327/full/v12/i25/4009.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i25.4009