Copyright
©2005 Baishideng Publishing Group Inc.
World J Gastroenterol. Oct 28, 2005; 11(40): 6366-6372
Published online Oct 28, 2005. doi: 10.3748/wjg.v11.i40.6366
Published online Oct 28, 2005. doi: 10.3748/wjg.v11.i40.6366
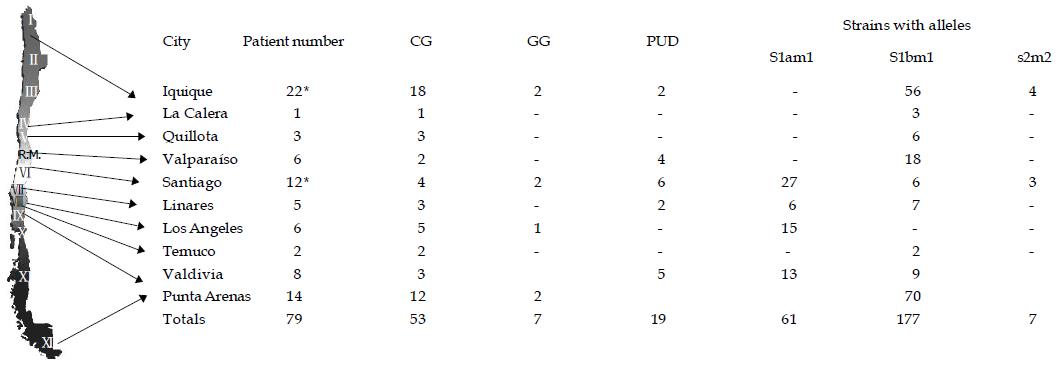
Figure 1 Chilean map and strain distribution according to the vacA alleles.
Map of Chile including the location of the cities where biopsies were taken. The corresponding vacA alleles, as well as the number of strains and patients are displayed (*). Two patients, one from Iquique and the other from Santiago, had mixed infections. Chronic Active Gastritis (CG), Gastropathy Group (GG), Peptic Ulcer Disease (PUD). Total amount of strains = 245.
Figure 2 Alignment of s and m alleles as vacA amino acid partial sequences.
A: Alignment of amino acid sequences corresponding to the amplified m1 vacA allele of Chilean strains CH-CTX1 and LA-12 with other representative m1 Indian sequences obtained from the GenBank; B: Alignment of amino acid sequences corresponding to the amplified m2 vacA allele and other representative m2 sequences obtained from the GenBank. The m2 allele from strain SA-64 was amplified using primers VA4F and VA4R and both strands were sequenced. The oligonucleotide sequence was translated (126 amino acids) and compared with other representative m2 allele sequences. These m2 sequences were obtained from GenBanK and corresponded to strains 95-54 from Italy, India 90, 97-203 from USA and V-296 from Taiwan; C: Alignment of amino acid sequences corresponding to Chilean strains having the s1a allele (CH-CTX1 and LA-12) and comparison with a s1a sequences from China. Sequences of Chilean s1b strains (IQ-61, QU-31 and QU-61) and comparison with other s1b sequences obtained from the GenBank from Argentinean strains AR-312 and AR-710; D: Alignment and comparison of amino acid sequences corresponding to the s2 sequence of the Chilean strain SA-64 and other sequences obtained from the GenBank of strains isolated in different locations. Sequences of the s2 vacA allele from different strains were aligned starting from amino acid position 30 in the vacA precursor sequence and the segment was fit with the ends defined by the Atherton primers for amplification of the s2 allele. ◆ Includes identical s2 sequences for strains: SA-64 from Chile, USA2781 from Arizona, USA, SS-1 from Australia, NA2010 and NA1986 from Colombia. ●Represents identical s2 sequences from strains NA1995, NA1992 and NA1685 from Colombia, India 3 and Alaska 5 from USA. Accession numbers are displayed in Materials and methods.
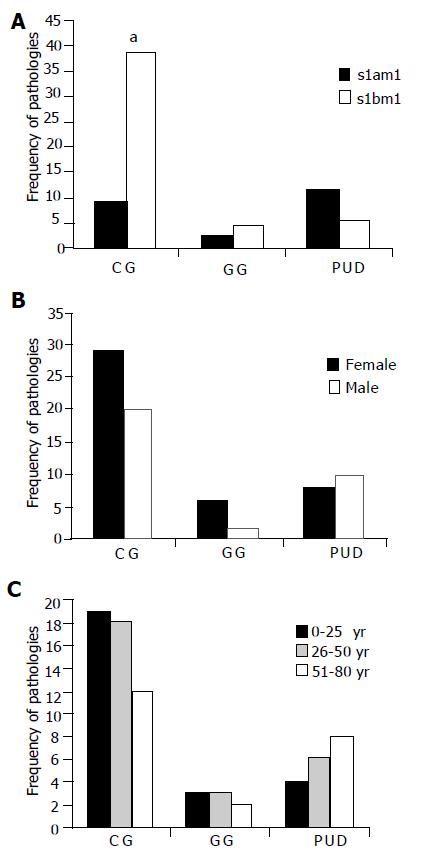
Figure 3 Analysis of gastropathies outcome frequencies respect to VacA.
genotype, gender and patient age; A: Relationship between pathologies and alleles. Minitab Release 12 PA.USA and χ2 test for categorical variables was used (P<0.05). P <0.05 was considered significant. The s2m2 allele was excluded of the statistics test due a low frequency (3%); B: Relationship between gastropathies and gender. F: female, M: male. A total of 43 females and 32 males were studied (P >0.5), four patients carrying strains with s2 m2 allele were excluded from this study; C: Relationship between gastropathies and patient age (P>0.5).
-
Citation: Díaz M, Valdivia A, Martínez P, Palacios J, Harris P, Novales J, Garrido E, Valderrama D, Shilling C, Kirberg A, Hebel E, Fierro J, Bravo R, Siegel F, Leon G, Klapp G, Venegas A.
Helicobacter pylori vacA s1a and s1b alleles from clinical isolates from different regions of Chile show a distinct geographic distribution. World J Gastroenterol 2005; 11(40): 6366-6372 - URL: https://www.wjgnet.com/1007-9327/full/v11/i40/6366.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i40.6366