Copyright
©The Author(s) 2004.
World J Gastroenterol. Nov 15, 2004; 10(22): 3284-3288
Published online Nov 15, 2004. doi: 10.3748/wjg.v10.i22.3284
Published online Nov 15, 2004. doi: 10.3748/wjg.v10.i22.3284
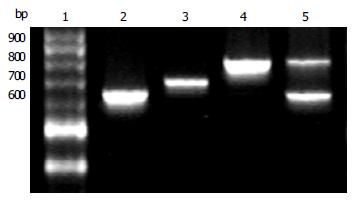
Figure 1 Amplified products of 3’ region of cagA gene by PCR (20 g/L agarose gel electrophoresis) Lane 1: 100 bp DNA ladder; Lane 2: subtype I (648 bp); Lane 3: subtype II (705 bp); Lane 4: subtype III (815 bp); Lane 5: from a patient with both subtypes I and III.
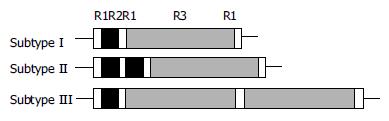
Figure 2 Primary structures of three subtypes of 3’ region of cagA gene R1: 15 bp repeat region; R2: 42 bp repeat region; R3: 147 bp repeat region.
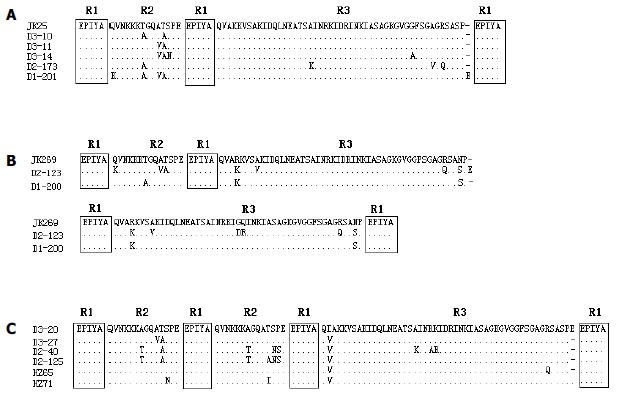
Figure 3 Alignment of amino acid sequences of subtypes of 3’ region of cagA gene.
A: Alignment of the deduced amino acid sequences of subtype I strains with the sequence of the corresponding region of a Japanese type A strain JK25 (GenBank accession number AF043487). Strain D2-173 was from a patient with chronic gastritis, and the remaining four strains were from patients with peptic ulcer. B: Alignment of the deduced amino acid sequences of subtype III strains with the sequence of the corresponding region of a Japanese type C strain JK269 (GenBank accession number AF043489). Strain D1-200 was from a patient with chronic gastritis, and strain D2-123 was from a patient with gastric ulcer. C: Alignment of the deduced amino acid sequences of six subtype II strains. Strain HZ65 and HZ71 were from patients with chronic gastritis, strain D2-40 was from a patient with gastric cancer, and the remaining three strains were from patients with peptic ulcer.
-
Citation: Tao R, Fang PC, Liu HY, Jiang YS, Chen J. A new subtype of 3’ region of
cagA gene inHelicobacter pylori strains isolated from Zhejiang Province in China. World J Gastroenterol 2004; 10(22): 3284-3288 - URL: https://www.wjgnet.com/1007-9327/full/v10/i22/3284.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i22.3284