Published online May 16, 2024. doi: 10.12998/wjcc.v12.i14.2370
Revised: February 11, 2024
Accepted: April 2, 2024
Published online: May 16, 2024
In observational studies, dietary intakes are associated with gastroesophageal re
To conduct a two-sample mendelian randomization (MR) analysis to determine whether those associations are causal.
To explore the relationship between dietary intake and the risk of GERD, we extracted appropriate single nucleotide polymorphisms from genome-wide asso
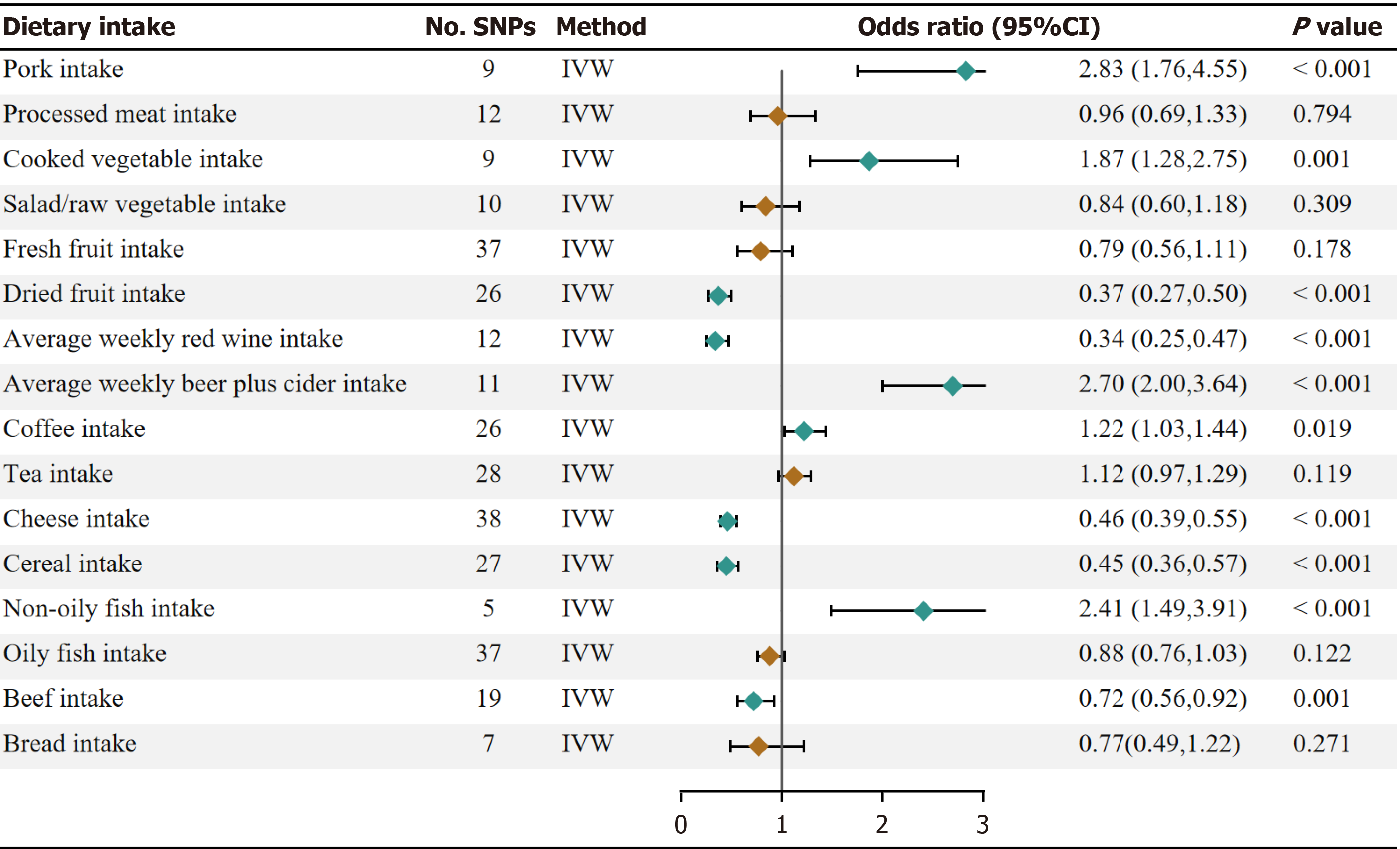
Our univariate Mendelian randomization (UVMR) results showed significant evidence that pork intake (OR, 2.83; 95%CI: 1.76-4.55; P = 1.84 × 10–5), beer intake (OR, 2.70, 95%CI: 2.00-3.64; P = 6.54 × 10–11), non-oily fish intake (OR, 2.41; 95%CI: 1.49-3.91; P = 3.59 × 10–4) have a protective effect on GERD. In addition, dried fruit intake (OR, 0.37; 95%CI: 0.27-0.50; 6.27 × 10–11), red wine intake (OR, 0.34; 95%CI: 0.25-0.47; P = 1.90 × 10-11), cheese intake (OR, 0.46; 95%CI: 0.39-0.55; P =3.73 × 10-19), bread intake (OR, 0.72; 95%CI: 0.56-0.92; P = 0.0009) and cereal intake (OR, 0.45; 95%CI: 0.36-0.57; P = 2.07 × 10-11) were negatively associated with the risk of GERD. There was a suggestive asso
This study provides MR evidence to support the causal relationship between a broad range of dietary intake and GERD, providing new insights for the treatment and prevention of GERD.
Core Tip: Through genetic prediction, this study demonstrated the protective effect of dried fruit, red wine, cheese, bread, and cereal intake against gastroesophageal reflux disease (GERD) and the detrimental effects of pig, beer, and non-oily fish intake. Furthermore, even after accounting for body mass index, major depressive disorder, smoking, and alcohol con
- Citation: Liu YX, Yang WT, Li Y. Different effects of 24 dietary intakes on gastroesophageal reflux disease: A mendelian randomization. World J Clin Cases 2024; 12(14): 2370-2381
- URL: https://www.wjgnet.com/2307-8960/full/v12/i14/2370.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i14.2370
Gastroesophageal reflux disease (GERD) refers to the flow of gastric contents back into the esophagus, causing discomfort and complications[1]. Meanwhile, GERD can progress to Barrett's esophagus and even increase the risk of esophageal adenocarcinoma[2]. It is estimated that about 20% of people in Western countries suffer from GERD[3]. The prevalence of GERD has gradually transitioned from the developed world to developing countries[4]. GERD patients in developing countries face a financial burden and discomfort due to deficient appropriate treatment[5]. As an easily accessible and modifiable factor, many researchers have begun to focus on the impact of diet on GERD. A cohort study has demon
Mendelian randomization (MR) is a powerful tool for epidemiological research; The central idea is to use genetic va
No MR studies are exploring the causal effect of multiple diets on GERD. We conducted a two sample MR study to examine the correlation between 24 dietary intake and GERD risk.
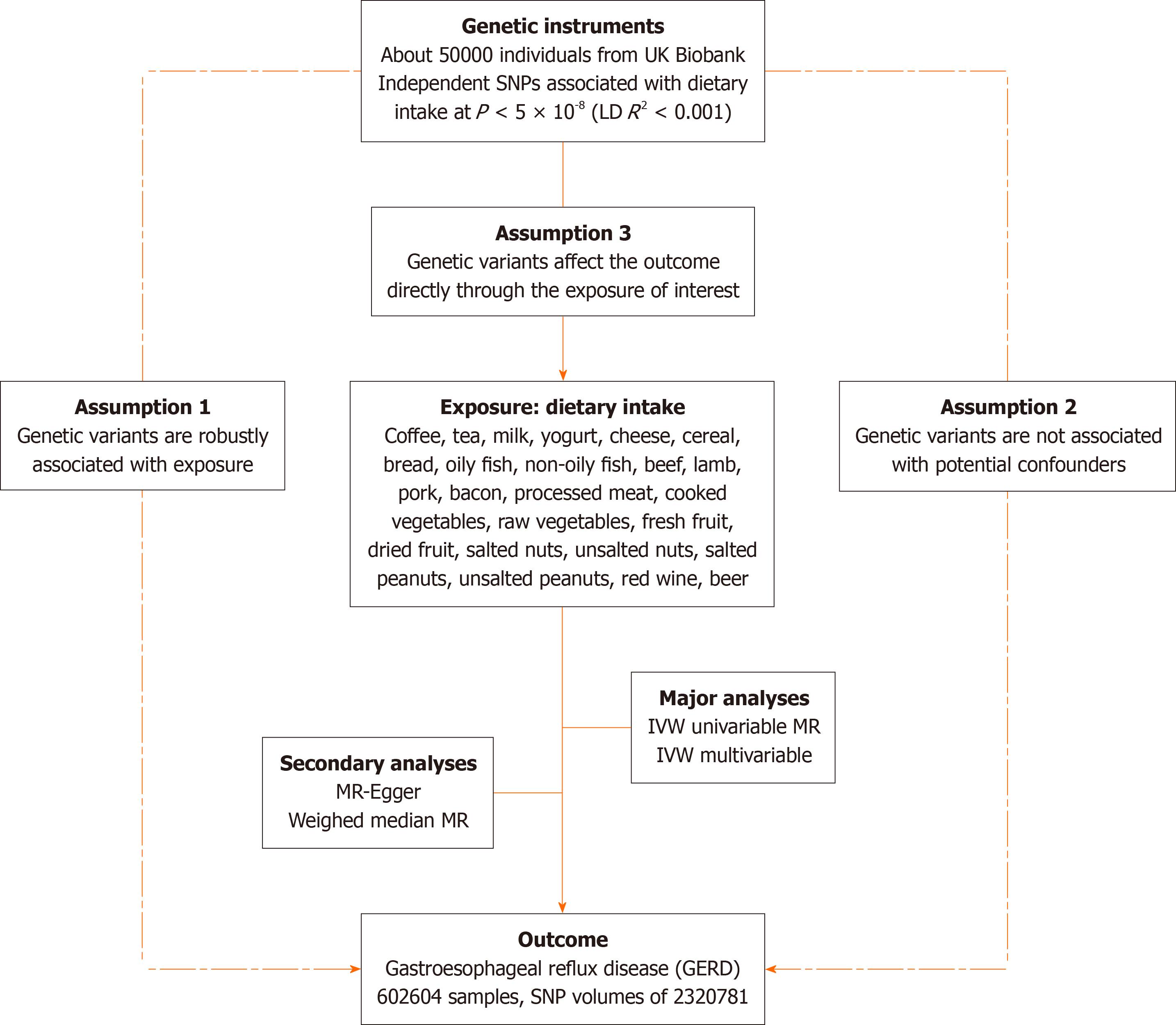
We evaluated the causal effects of 24 dietary incomes on GERD using two-sample Mendelian randomization. Then, we used multivariable MR (MVMR) to adjust for risk factors that could affect GERD occurrence. Our MR study is based on three hypotheses: Genetic variants are closely associated with the exposure of interest, not causally related to the outcome but only through the exposure, and not confounded by other variables[15]. An overview of the principles, design, and procedures of our MR study is shown in Figure 1.
Genetic variations of 24 dietary intakes were collected from participants of the UK Biobank cohort. Related exposure included coffee, tea, milk, yogurt, cheese, cereal, bread, oily fish, non-oily fish, beef, lamb, pork, bacon, processed meat, cooked vegetables, raw vegetables, fresh fruit, dried fruit, salted nuts, unsalted nuts, salted peanuts, unsalted peanuts, red wine, and beer. Genetic data for gastroesophageal reflux disease was also obtained from the genome-wide association study (GWAS) catalog database with single nucleotide polymorphisms (SNP) volumes of 2320781[16]. Furthermore, we identified variables commonly associated with esophageal disorders: body mass index (BMI)[17], major depressive dis
| Dietary intake | R2 | F-statistic | SNPs | IVW | WM | MR-egger | |||
| OR (95%CI) | P value | OR (95%CI) | P value | OR (95%CI) | P value | ||||
| Pork intake | 0.0004 | 20.499 | 9 | 2.83 (1.76, 4.55) | 1.84E-05 | 3.60 (2.14, 6.07) | 1.52E-06 | 49.55 (1.55, 1579.54) | 0.063 |
| Bacon intake | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Processed meat intake | 0.0014 | 40.506 | 12 | 0.96 (0.69, 1.33) | 0.794 | 1.12 (0.78, 1.59) | 0.544 | 0.19 (0.01, 3.67) | 0.296 |
| Cooked vegetable intake | 0.0003 | 10.983 | 9 | 1.87 (1.28, 2.75) | 0.001 | 1.56 (0.95, 2.55) | 0.081 | 0.71 (0.01, 64.25) | 0.885 |
| Salad/raw vegetable intake | 0.0003 | 18.628 | 10 | 0.84 (0.60, 1.18) | 0.309 | 0.90 (0.57, 1.42) | 0.639 | 2.39 (0.42, 13.44) | 0.352 |
| Fresh fruit intake | 0.0008 | 18.132 | 37 | 0.79 (0.56, 1.11) | 0.178 | 0.87 (0.60, 1.27) | 0.472 | 1.65 (0.46, 5.88) | 0.443 |
| Dried fruit intake | 0.0009 | 12.062 | 26 | 0.37 (0.27, 0.50) | 6.27E-11 | 0.44 (0.30, 0.61) | 9.00E-07 | 0.13 (0.02, 0.86) | 0.045 |
| Salted nuts intake | NA | NA | 1 | NA | NA | NA | NA | NA | NA |
| Unsalted nuts intake | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Salted peanuts intake | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Unsalted peanuts intake | NA | NA | 1 | NA | NA | NA | NA | NA | NA |
| Average weekly red wine intake | 0.0007 | 15.584 | 12 | 0.34 (0.25, 0.47) | 1.90E-11 | 0.33 (0.24, 0.47) | 7.03E-10 | 0.35 (0.04, 3.37) | 0.388 |
| Average weekly beer plus cider intake | 0.0005 | 11.283 | 11 | 2.70 (2.00, 3.64) | 6.54E-11 | 2.59 (1.75, 3.83) | 1.82E-06 | 5.19 (0.73, 36.97) | 0.134 |
| Coffee intake | 0.0017 | 23.483 | 26 | 1.22 (1.03, 1.44) | 0.019 | 1.28 (1.06, 1.56) | 0.010 | 1.43 (1.05, 1.94) | 0.034 |
| Tea intake | 0.0025 | 33.827 | 28 | 1.12 (0.97, 1.29) | 0.119 | 1.23 (1.04, 1.45) | 0.014 | 1.32 (0.97, 1.80) | 0.086 |
| Milk intake | NA | NA | 2 | NA | NA | NA | NA | NA | NA |
| Yogurt intake | NA | NA | 1 | NA | NA | NA | NA | NA | NA |
| Cheese intake | 0.0020 | 21.543 | 38 | 0.46 (0.39, 0.55) | 3.73E-19 | 0.57 (0.47, 0.69) | 8.65E-09 | 0.83 (0.33, 2.13) | 0.704 |
| Cereal intake | 0.0012 | 16.373 | 27 | 0.45 (0.36, 0.57) | 2.07E-11 | 0.49 (0.38, 0.63) | 4.36E-08 | 0.58 (0.20, 1.64) | 0.314 |
| Non-oily fish intake | 0.0002 | 13.416 | 5 | 2.41 (1.49, 3.91) | < 0.001 | 1.96 (1.06, 3.62) | 0.033 | 13.70 (0.11, 1761.13) | 0.368 |
| Oily fish intake | 0.0020 | 19.800 | 37 | 0.88 (0.76, 1.03) | 0.122 | 0.89 (0.74, 1.08) | 0.244 | 0.64 (0.32, 1.30) | 0.227 |
| Lamb intake | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Beef intake | 0.0004 | 15.600 | 19 | 0.72 (0.56, 0.92) | 0.001 | 0.80 (0.61, 1.05) | 0.108 | 0.69 (0.25, 1.86) | 0.470 |
| Bread intake | 0.0010 | 20.202 | 7 | 0.77 (0.49, 1.22) | 0.271 | 0.57 (0.36, 0.91) | 0.018 | 0.03 (0.00, 0.25) | 0.022 |
First, SNPs with significant association with dietary intake (P < 5.0 × 10-8) were selected. A parameter R2 threshold of 0.001 and a kilobase pair (kb) of 10000 were set to exclude interference from linkage disequilibrium (LD)[20]. Then, The SNPs were obtained and isolated from the outcome data, and the SNPs significantly associated with the outcomes (P < 1×10−5) were excluded[21]. If any SNPs were not found in the outcome datasets, proxies with LD R2 > 0.8 were used[22]. How
Three methods were used for MR analysis: inverse variance weighted analysis (IVW), MR egger, and weighted median. The IVW approach integrates the Wald ratio estimated for each SNP through meta-analysis[27]. IVW method was used as the primary statistical method, which is divided into two models: fixed effect (exposure constructed by ≥ 3 SNPs) and random effect (exposure constructed by < 3 SNPs)[27]. We prioritize using random effect-IVW, which assumes that MR estimates obtained for different SNPs conform to a normal distribution. This assumption is more reasonable and is somewhat tolerant of heterogeneity[28]. Assuming that > 50% of the weights come from effective SNPs, the weighted median (WM) method can provide consistent estimates. It has lower statistical efficacy than the IVW method[29]. The MR-Egger method is the most tolerant of horizontal pleiotropy, allowing all SNPs to fail to satisfy the three MR hypo
The MRPRESSO method is a useful tool to evaluate horizontal pleiotropy. It consists of three components: Firstly, the MR-PRESSO global test is used to detect the presence of horizontal pleiotropy. Secondly, the MR-PRESSO outlier test is utilized to remove any abnormal SNPs (outliers) and estimate the corrected outcome, which eliminates horizontal pleio
The study used the 95% confidence interval (CI) of the odds ratio (OR) to evaluate the impact of dietary intakes on GERD. P < 0.05 was considered suggestive; Significant associations required P < 0.002 (= 0.05/24) by Bonferroni cor
Supplementary Tables 2-17 show SNPs associated with 24 dietary intake and GERD. The total F-value of the intake of cooked vegetables, salad/raw vegetables, and fresh fruits is less than 10, indicating a weak instrumental bias among these three variables. Therefore, it is believed that there is no causal relationship between them and GERD. The F statistics for the rest of the phenotypes was > 10, indicating a small probability of weak instrument variable bias. Furthermore, we applied Steiger filtering to determine the accurate direction of inference.
Higher genetically predicted pork intake, beer intake, and non-oily fish intake were associated with an increased risk of GERD. The OR of GERD was 2.83 (95% confidence interval (CI), 1.76, 4.55; P = 1.84 × 10–5) for one standard deviation (SD) increase in pork intake, 2.70 (95%CI: 2.00-3.64; P = 6.54 × 10–11) for a one-unit increase in log-transformed OR of beer intake, and 2.41 (95%CI: 1.49-3.91; P = 3.59 × 10–4) for one SD increase in non-oily fish intake. In addition, dried fruit intake (OR 0.37; 95%CI: 0.27-0.50; 6.27 × 10–11), red wine intake (OR 0.34; 95%CI: 0.25-0.47; P = 1.90 × 10-11), cheese intake (OR 0.46; 95%CI: 0.39-0.55; P = 3.73 × 10-19), bread intake (OR, 0.72; 95%CI: 0.56-0.92; P = 0.0009), and cereal intake (OR 0.45; 95%CI: 0.36-0.57; P = 2.07 × 10-11) were negatively associated with the risk of GERD. There was a suggestive asso
The estimates from other MR methods, including WM and MR-Egger, consistently supported the causal inferences. Furthermore, there is no causal relationship between other dietary intake and GERD. In sensitivity analyses, the MR-PRESSO Distortion Test found outliers in the 16 dietary intakes (Supplementary Table 2-17). After excluding outliers, the nominal association between dietary intakes and GERD remained consistent. An analysis of the relationship between beef intake and GERD showed evidence of horizontal pleiotropy (P for MR-Egger intercept < 0.05) (Table 2). Leave-one-out analysis further supported that any single SNP did not drive the causalities (Supplementary Figures H1-16). Additionally, the funnel plot results indicated a symmetrical distribution of causal association effects when using SNPs individually as instrumental variables, and no potential bias was detected (Supplementary Figures S1-16). The forest plot also demon
| Dietary intake | No. SNPs | Heterogeneity | Pleiotropy | ||||||
| Q-MR Egger | Q-IVW | P-MR Egger | P-IVW | Intercept | SE | P value | MRPRESSO global test P | ||
| Pork intake | 9 | 10.80 | 14.92 | 0.148 | 0.061 | -0.028 | 0.018 | 0.146 | 0.091 |
| Bacon intake | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Processed meat intake | 12 | 22.74 | 25.39 | 0.012 | 0.008 | 0.023 | 0.022 | 0.306 | 0.01 |
| Cooked vegetable intake | 9 | 9.61 | 9.86 | 0.212 | 0.275 | 0.036 | 0.032 | 0.242 | 0.314 |
| Salad / raw vegetable intake | 10 | 7.800 | 9.26 | 0.453 | 0.414 | -0.011 | 0.009 | 0.262 | 0.39 |
| Fresh fruit intake | 37 | 119.23 | 124.00 | 4.05E-11 | 1.35E-11 | -0.007 | 0.006 | 0.245 | < 0.001 |
| Dried fruit intake | 26 | 66.08 | 69.47 | 8.43E-06 | 4.61E-06 | 0.012 | 0.011 | 0.278 | < 0.001 |
| Salted nuts intake | 1 | NA | NA | NA | NA | NA | NA | NA | NA |
| Unsalted nuts intake | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Salted peanuts intake | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Unsalted peanuts intake | 1 | NA | NA | NA | NA | NA | NA | NA | NA |
| Average weekly red wine intake | 12 | 24.28 | 24.28 | 0.007 | 0.012 | -0.001 | 0.016 | 0.974 | 0.043 |
| Average weekly beer plus cider intake | 11 | 12.74 | 13.36 | 0.175 | 0.204 | -0.008 | 0.012 | 0.525 | 0.32 |
| Coffee intake | 26 | 43.08 | 45.59 | 0.010 | 0.007 | -0.003 | 0.003 | 0.249 | 0.008 |
| Tea intake | 28 | 52.56 | 55.47 | 0.002 | 0.001 | -0.004 | 0.003 | 0.241 | 0.002 |
| Milk intake | 2 | NA | NA | NA | NA | NA | NA | NA | NA |
| Yogurt intake | 1 | NA | NA | NA | NA | NA | NA | NA | NA |
| Cheese intake | 38 | 80.85 | 84.39 | 2.70E-05 | 1.45E-05 | -0.009 | 0.007 | 0.217 | < 0.001 |
| Cereal intake | 27 | 60.22 | 60.77 | 9.74E-05 | 1.32E-04 | -0.004 | 0.007 | 0.638 | 0.003 |
| Non-oily fish intake | 5 | 4.17 | 4.86 | 0.244 | 0.302 | -0.018 | 0.026 | 0.531 | 0.376 |
| Oily fish intake | 37 | 56.19 | 57.51 | 0.013 | 0.013 | 0.004 | 0.005 | 0.369 | 0.015 |
| Lamb intake | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Beef intake | 19 | 4.43 | 11.58 | 0.619 | 0.115 | 0.031 | 0.012 | 0.037 | 0.138 |
| Bread intake | 7 | 42.02 | 42.04 | 0.001 | 0.001 | 0.001 | 0.007 | 0.928 | 0.002 |
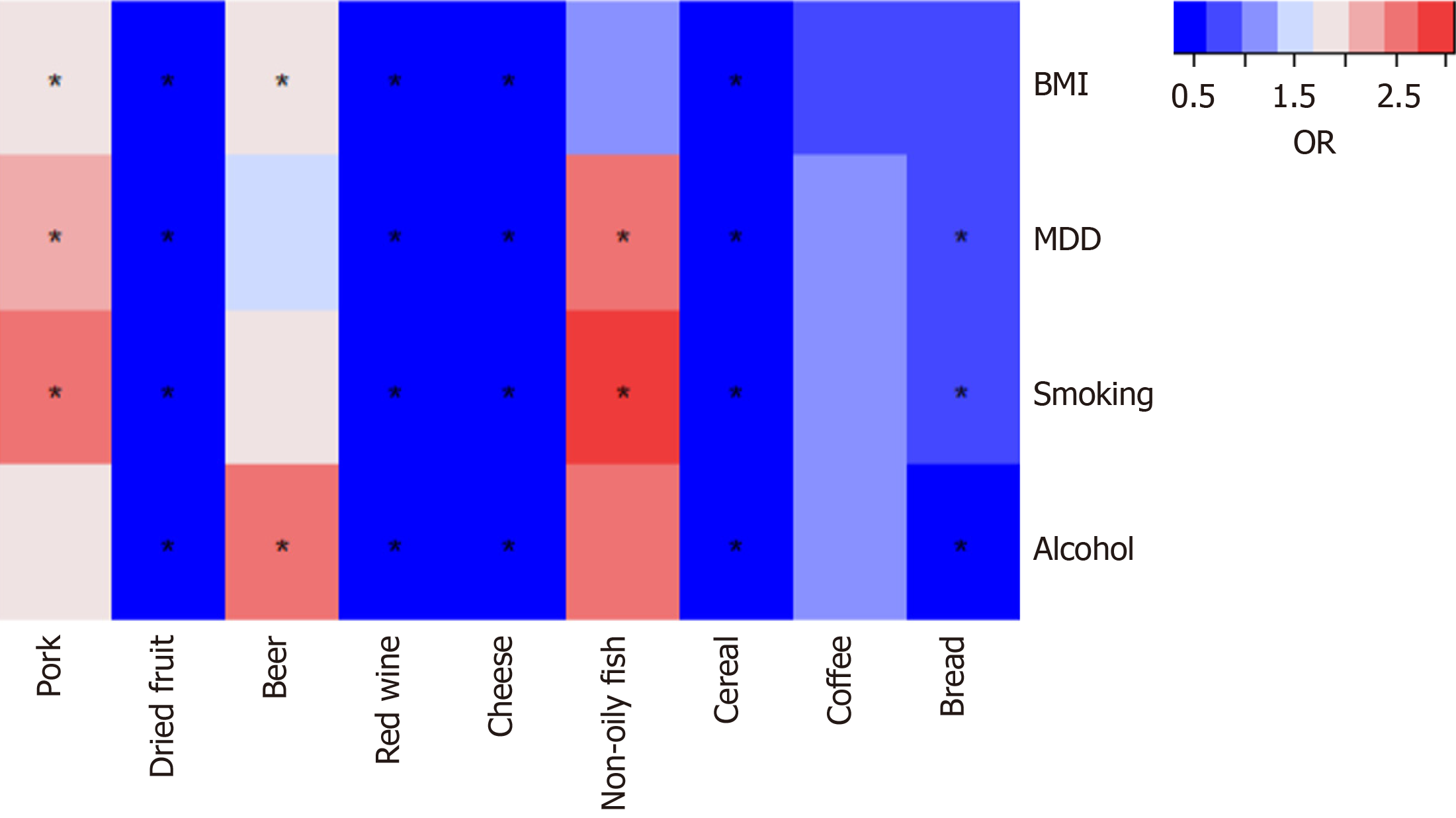
To determine whether the nine dietary intake directly or through common GERD risk factors affect GERD risk, we con
This MR study found that higher genetically predicted pork intake, beer intake, and non-oily fish intake were associated with an increased risk of GERD. Moreover, we found that dried fruit, red wine, cheese, bread, and cereal have a pro
For dried fruit and GERD, a retrospective study from Maekita T found that daily intake of dried Japanese apricots helped improve GERD symptoms[34]. However, an animal model study found that consuming dried fruits had no effect on the cellular antioxidant status in rats with reflux-induced esophagitis[35]. Our study found a significant protective effect of dried fruits against GERD after adjusting for BMI, MDD, smoking, and alcohol drinking. This strongly indicates that this protective effect is at least unrelated to the common risk factors of GERD. Dried fruits contain a variety of macronutrients, micronutrients, and health-promoting bioactive. These compounds exhibit antioxidant and free radical scavenging activities, which help improve digestive tract disorders[36]. A meta-analysis suggests that dried fruits have preventive value against certain cancers, particularly cancers of the digestive system[37]. Further research is needed on how dried fruits can reduce the increased risk of GERD.
Between alcohol consumption and GERD, the MR study by Yuan et al[38] found that genetic prediction of alcohol con
Fermented dairy products are known to be nutritious, high in probiotics, and rich in calcium-quality proteins, bioactive molecules, vitamins, and other ingredients[42]. Their availability can be increased due to the fermentation process[43]. A retrospective study suggests high consumption of milk products and dietary fat is associated with severe GERD symp
Dietary fiber, particularly from cereal sources, has been found to be linked to a lower risk of adenocarcinoma in the esophagus and gastric cardia[47]. A case-control study from M Nilsson showed that the risk of reflux was significantly reduced as the amount of dietary fiber increased[48]. This is highly consistent with our findings. In addition, cereal intake played an independent and significant role after excluding the effects of risk factors. The biological mechanism under
Our study found pork intake increased GERD risk. This is consistent with the results of several observational studies[51,52]. Further MVMR analysis indicated that the harmful effect of pork intake on GERD might be driven by alcohol assumption. Red meat is rich in hemoglobin and iron, which can catalytically oxidize and cause oxidative stress damage to the body[53]. Then, this can cause wear on the esophageal sphincter and exacerbate reflux. Similar to pork intake, our study found that non-oily fish intake enhances the risk of GERD development. A cross-sectional study in China found that the prevalence of GERD was increased by excessive non-oily fish intake[54]. Additionally, BMI and alcoholic drink
There are several observational studies on the effects of coffee on GERD, and their evidence results are inconsistent[55-57]. Hence, there is a lack of high-level evidence to confirm the association. Our MR study suggested that coffee intake has a suggestive association with GERD before adjusting for four risk factors. However, after adjusting for all four ele
One of the advantages of this study is that it comprehensively characterizes the relationship between dietary intakes and GERD through MR analysis. Second, our analysis is superior to previous studies as we used pooled data from GWAS with larger sample sizes and more SNPs, avoiding biases such as unobserved confounding, misclassification, and reverse causation. Third, we also adjusted for the effect of some risk factors for GERD, further validating the second hypothesis of MR.
This study has some noticeable drawbacks. Firstly, horizontal pleiotropy is a major limitation in MR design, where SNPs affect outcomes through alternative pathways rather than exposure[31]. We used the MR-Egger intercept and MRPRESSO global test to detect pleiotropy. After excluding outliers, there was still horizontal pleiotropy for several phenotypes in the MRPRESSO global test. However, we found no evidence of horizontal pleiotropy in the MR-Egger analysis, which is consistent with the results of several sensitivity analyses. Secondly, this study only covered European populations, which may limit its applicability to other ethnic groups. Finally, we found different causal effect estimates for the MR-Egger and other MR methods. Due to its calculation of horizontal pleiotropy, it has weaker statistical efficacy than other MR methods. Our primary approach is to rely on the findings from the IVW method.
To our knowledge, there have been numerous MR studies investigating the risk factors and protective factors of GERD[59-61]. However, there are few studies on the intake of meat, staple foods, fruits, vegetables, and beverages. GERD has a severe impact on the quality of life of patients and lacks an effective treatment. Our conclusions can help clinicians to educate patients about their health and to develop suitable recipes for patients with GERD. For GERD patients, dietary changes can be made to alleviate reflux symptoms and reduce financial burdens.
This study revealed the protective effects of dry fruit intake, red wine intake, cheese intake, bread intake, and grain intake on GERD through genetic prediction, as well as the harmful effects of pork intake, beer intake, and non-oily fish intake on GERD. Furthermore, the effect of genetically predicted dried fruit, red wine, cheese, and cereal on GERD remained after adjusting for BMI, MDD, smoking, and alcohol drinking. Higher genetically forecasted coffee intake was suggestively associated with GERD. However, after adjusting for all four factors, there was no longer a suggestive association between coffee intake and GERD. This study also found that tea intake, milk intake, yogurt intake, oily fish intake, beef intake, lamb intake, bacon intake, processed meat intake, cooked vegetable intake, raw vegetable intake, fresh fruit intake, salted nuts intake, unsalted nuts intake, salted peanuts intake, unsalted peanuts intake were not associated with GERD.
We thank the contributors of the original GWAS datasets.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Oprea VD, Romania S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Maret-Ouda J, Markar SR, Lagergren J. Gastroesophageal Reflux Disease: A Review. JAMA. 2020;324:2536-2547. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Mittal R, Vaezi MF. Esophageal Motility Disorders and Gastroesophageal Reflux Disease. N Engl J Med. 2020;383:1961-1972. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Richter JE, Rubenstein JH. Presentation and Epidemiology of Gastroesophageal Reflux Disease. Gastroenterology. 2018;154:267-276. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | He J, Ma X, Zhao Y, Wang R, Yan X, Yan H, Yin P, Kang X, Fang J, Hao Y, Li Q, Dent J, Sung JJ, Zou D, Wallander MA, Johansson S, Liu W, Li Z. A population-based survey of the epidemiology of symptom-defined gastroesophageal reflux disease: the Systematic Investigation of Gastrointestinal Diseases in China. BMC Gastroenterol. 2010;10:94. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Mikami DJ, Murayama KM. Physiology and pathogenesis of gastroesophageal reflux disease. Surg Clin North Am. 2015;95:515-525. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Mehta RS, Nguyen LH, Ma W, Staller K, Song M, Chan AT. Association of Diet and Lifestyle With the Risk of Gastroesophageal Reflux Disease Symptoms in US Women. JAMA Intern Med. 2021;181:552-554. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Kubo A, Block G, Quesenberry CP Jr, Buffler P, Corley DA. Dietary guideline adherence for gastroesophageal reflux disease. BMC Gastroenterol. 2014;14:144. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Kim JS, Kim BW. Are Diet and Micronutrients Effective in Treating Gastroesophageal Reflux Disease Especially in Women? J Neurogastroenterol Motil. 2019;25:1-2. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Rivière P, Vauquelin B, Rolland E, Melchior C, Roman S, Bruley des Varannes S, Mion F, Gourcerol G, Sacher-Huvelin S, Zerbib F. Low FODMAPs diet or usual dietary advice for the treatment of refractory gastroesophageal reflux disease: An open-labeled randomized trial. Neurogastroenterol Motil. 2021;33:e14181. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Zhang M, Hou ZK, Huang ZB, Chen XL, Liu FB. Dietary and Lifestyle Factors Related to Gastroesophageal Reflux Disease: A Systematic Review. Ther Clin Risk Manag. 2021;17:305-323. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133-1163. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Zhu X, Li X, Xu R, Wang T. An iterative approach to detect pleiotropy and perform Mendelian Randomization analysis using GWAS summary statistics. Bioinformatics. 2021;37:1390-1400. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | O'Donnell CJ, Sabatine MS. Opportunities and Challenges in Mendelian Randomization Studies to Guide Trial Design. JAMA Cardiol. 2018;3:967. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA. 2017;318:1925-1926. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Ong JS, An J, Han X, Law MH, Nandakumar P; 23andMe Research team; Esophageal cancer consortium, Schumacher J, Gockel I, Bohmer A, Jankowski J, Palles C, Olsen CM, Neale RE, Fitzgerald R, Thrift AP, Vaughan TL, Buas MF, Hinds DA, Gharahkhani P, Kendall BJ, MacGregor S. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut. 2022;71:1053-1061. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Mägi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman ÅK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stančáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Ärnlöv J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Blüher M, Böhringer S, Bonnycastle LL, Böttcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen YI, Clarke R, Daw EW, de Craen AJM, Delgado G, Dimitriou M, Doney ASF, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Gräßler J, Grönberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson Å, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lo KS, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PKE, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Müller G, Müller-Nurasyid M, Musk AW, Nagaraja R, Nöthen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Smith AV, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q; LifeLines Cohort Study, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gådin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JRB, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van 't Hooft FM, Vinkhuyzen AAE, Westra HJ, Zheng W, Zondervan KT; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium, Heath AC, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hyppönen E, Illig T, Jacobs KB, Jarvelin MR, Jöckel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJP, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Marchand LL, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PAF, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PEH, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tönjes A, Trégouët DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Völker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CNA, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O'Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJF, Speliotes EK. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197-206. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu SA, Bækvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschøn HN, Bybjerg-Grauholm J, Cai N, Castelao E, Christensen JH, Clarke TK, Coleman JIR, Colodro-Conde L, Couvy-Duchesne B, Craddock N, Crawford GE, Crowley CA, Dashti HS, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV, Dunn EC, Eley TC, Eriksson N, Escott-Price V, Kiadeh FHF, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Giusti-Rodríguez P, Goes FS, Gordon SD, Grove J, Hall LS, Hannon E, Hansen CS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga JJ, Hougaard DM, Hu M, Hyde CL, Ising M, Jansen R, Jin F, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Lane JM, Li Y, Lind PA, Liu X, Lu L, MacIntyre DJ, MacKinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, McGrath P, McGuffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mill J, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B, Nivard MG, Nyholt DR, O'Reilly PF, Oskarsson H, Owen MJ, Painter JN, Pedersen CB, Pedersen MG, Peterson RE, Pettersson E, Peyrot WJ, Pistis G, Posthuma D, Purcell SM, Quiroz JA, Qvist P, Rice JP, Riley BP, Rivera M, Saeed Mirza S, Saxena R, Schoevers R, Schulte EC, Shen L, Shi J, Shyn SI, Sigurdsson E, Sinnamon GBC, Smit JH, Smith DJ, Stefansson H, Steinberg S, Stockmeier CA, Streit F, Strohmaier J, Tansey KE, Teismann H, Teumer A, Thompson W, Thomson PA, Thorgeirsson TE, Tian C, Traylor M, Treutlein J, Trubetskoy V, Uitterlinden AG, Umbricht D, Van der Auwera S, van Hemert AM, Viktorin A, Visscher PM, Wang Y, Webb BT, Weinsheimer SM, Wellmann J, Willemsen G, Witt SH, Wu Y, Xi HS, Yang J, Zhang F; eQTLGen; 23andMe, Arolt V, Baune BT, Berger K, Boomsma DI, Cichon S, Dannlowski U, de Geus ECJ, DePaulo JR, Domenici E, Domschke K, Esko T, Grabe HJ, Hamilton SP, Hayward C, Heath AC, Hinds DA, Kendler KS, Kloiber S, Lewis G, Li QS, Lucae S, Madden PFA, Magnusson PK, Martin NG, McIntosh AM, Metspalu A, Mors O, Mortensen PB, Müller-Myhsok B, Nordentoft M, Nöthen MM, O'Donovan MC, Paciga SA, Pedersen NL, Penninx BWJH, Perlis RH, Porteous DJ, Potash JB, Preisig M, Rietschel M, Schaefer C, Schulze TG, Smoller JW, Stefansson K, Tiemeier H, Uher R, Völzke H, Weissman MM, Werge T, Winslow AR, Lewis CM, Levinson DF, Breen G, Børglum AD, Sullivan PF; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668-681. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X; 23andMe Research Team; HUNT All-In Psychiatry, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga JJ, Huang H, Jang SK, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237-244. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Li M, Lin J, Liang S, Chen Z, Bai Y, Long X, Huang S, Mo Z. The role of age at menarche and age at menopause in Alzheimer's disease: evidence from a bidirectional mendelian randomization study. Aging (Albany NY). 2021;13:19722-19749. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Shen J, Zhou H, Liu J, Zhang Y, Zhou T, Yang Y, Fang W, Huang Y, Zhang L. A modifiable risk factors atlas of lung cancer: A Mendelian randomization study. Cancer Med. 2021;10:4587-4603. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Cui Z, Hou G, Meng X, Feng H, He B, Tian Y. Bidirectional Causal Associations Between Inflammatory Bowel Disease and Ankylosing Spondylitis: A Two-Sample Mendelian Randomization Analysis. Front Genet. 2020;11:587876. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45:1717-1726. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251-260. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48:713-727. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Zhang J, Taylor EW, Bennett K, Saad R, Rayman MP. Association between regional selenium status and reported outcome of COVID-19 cases in China. Am J Clin Nutr. 2020;111:1297-1299. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology. 2017;28:30-42. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Yin KJ, Huang JX, Wang P, Yang XK, Tao SS, Li HM, Ni J, Pan HF. No Genetic Causal Association Between Periodontitis and Arthritis: A Bidirectional Two-Sample Mendelian Randomization Analysis. Front Immunol. 2022;13:808832. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-314. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Slob EAW, Groenen PJF, Thurik AR, Rietveld CA. A note on the use of Egger regression in Mendelian randomization studies. Int J Epidemiol. 2017;46:2094-2097. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-698. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Magnus MC, Guyatt AL, Lawn RB, Wyss AB, Trajanoska K, Küpers LK, Rivadeneira F, Tobin MD, London SJ, Lawlor DA, Millard LAC, Fraser A. Identifying potential causal effects of age at menarche: a Mendelian randomization phenome-wide association study. BMC Med. 2020;18:71. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Millard LAC, Munafò MR, Tilling K, Wootton RE, Davey Smith G. MR-pheWAS with stratification and interaction: Searching for the causal effects of smoking heaviness identified an effect on facial aging. PLoS Genet. 2019;15:e1008353. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Maekita T, Kato J, Enomoto S, Yoshida T, Utsunomiya H, Hayashi H, Hanamitsu T, Inoue I, Maeda Y, Moribata K, Muraki Y, Shingaki N, Deguchi H, Ueda K, Iguchi M, Tamai H, Ichinose M. Japanese apricot improves symptoms of gastrointestinal dysmotility associated with gastroesophageal reflux disease. World J Gastroenterol. 2015;21:8170-8177. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Aiyer HS, Li Y, Liu QH, Reuter N, Martin RC. Dietary freeze-dried black raspberry's effect on cellular antioxidant status during reflux-induced esophagitis in rats. Nutrition. 2011;27:182-187. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Alasalvar C, Salvadó JS, Ros E. Bioactives and health benefits of nuts and dried fruits. Food Chem. 2020;314:126192. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Mossine VV, Mawhinney TP, Giovannucci EL. Dried Fruit Intake and Cancer: A Systematic Review of Observational Studies. Adv Nutr. 2020;11:237-250. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Yuan S, Larsson SC. Adiposity, diabetes, lifestyle factors and risk of gastroesophageal reflux disease: a Mendelian randomization study. Eur J Epidemiol. 2022;37:747-754. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Pehl C, Pfeiffer A, Wendl B, Kaess H. Different effects of white and red wine on lower esophageal sphincter pressure and gastroesophageal reflux. Scand J Gastroenterol. 1998;33:118-122. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Pehl C, Wendl B, Pfeiffer A. White wine and beer induce gastro-oesophageal reflux in patients with reflux disease. Aliment Pharmacol Ther. 2006;23:1581-1586. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | Seidl H, Gundling F, Schepp W, Schmidt T, Pehl C. Effect of low-proof alcoholic beverages on duodenogastro-esophageal reflux in health and GERD. Neurogastroenterol Motil. 2011;23:145-150, e29. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Savaiano DA, Hutkins RW. Yogurt, cultured fermented milk, and health: a systematic review. Nutr Rev. 2021;79:599-614. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Sethi S, Richter JE. Diet and gastroesophageal reflux disease: role in pathogenesis and management. Curr Opin Gastroenterol. 2017;33:107-111. [PubMed] [DOI] [Cited in This Article: ] |
| 44. | Erdman KA, Jones KW, Madden RF, Gammack N, Parnell JA. Dietary Patterns in Runners with Gastrointestinal Disorders. Nutrients. 2021;13. [PubMed] [DOI] [Cited in This Article: ] |
| 45. | Fernando I, Schmidt KA, Cromer G, Burhans MS, Kuzma JN, Hagman DK, Utzschneider KM, Holte S, Kraft J, Vaughan TL, Kratz M. The impact of low-fat and full-fat dairy foods on symptoms of gastroesophageal reflux disease: an exploratory analysis based on a randomized controlled trial. Eur J Nutr. 2022;61:2815-2823. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Liang Z, Song X, Hu J, Wu R, Li P, Dong Z, Liang L, Wang J. Fermented Dairy Food Intake and Risk of Colorectal Cancer: A Systematic Review and Meta-Analysis. Front Oncol. 2022;12:812679. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Roth J, Mobarhan S. Preventive role of dietary fiber in gastric cardia cancers. Nutr Rev. 2001;59:372-374. [PubMed] [DOI] [Cited in This Article: ] |
| 48. | Nilsson M, Johnsen R, Ye W, Hveem K, Lagergren J. Lifestyle related risk factors in the aetiology of gastro-oesophageal reflux. Gut. 2004;53:1730-1735. [PubMed] [DOI] [Cited in This Article: ] |
| 49. | Pera M, Manterola C, Vidal O, Grande L. Epidemiology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:151-159. [PubMed] [DOI] [Cited in This Article: ] |
| 50. | El-Serag HB, Satia JA, Rabeneck L. Dietary intake and the risk of gastro-oesophageal reflux disease: a cross sectional study in volunteers. Gut. 2005;54:11-17. [PubMed] [DOI] [Cited in This Article: ] |
| 51. | Ness-Jensen E, Hveem K, El-Serag H, Lagergren J. Lifestyle Intervention in Gastroesophageal Reflux Disease. Clin Gastroenterol Hepatol. 2016;14:175-82.e1. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Caselli M, Lo Cascio N, Rabitti S, Eusebi LH, Zeni E, Soavi C, Cassol F, Zuliani G, Zagari RM. Pattern of food intolerance in patients with gastro-esophageal reflux symptoms. Minerva Med. 2017;108:496-501. [PubMed] [DOI] [Cited in This Article: ] |
| 53. | Huang Y, Cao D, Chen Z, Chen B, Li J, Guo J, Dong Q, Liu L, Wei Q. Red and processed meat consumption and cancer outcomes: Umbrella review. Food Chem. 2021;356:129697. [PubMed] [DOI] [Cited in This Article: ] |
| 54. | Gong Y, Zeng Q, Yan Y, Han C, Zheng Y. Association between Lifestyle and Gastroesophageal Reflux Disease Questionnaire Scores: A Cross-Sectional Study of 37 442 Chinese Adults. Gastroenterol Res Pract. 2019;2019:5753813. [PubMed] [DOI] [Cited in This Article: ] |
| 55. | Mehta RS, Song M, Staller K, Chan AT. Association Between Beverage Intake and Incidence of Gastroesophageal Reflux Symptoms. Clin Gastroenterol Hepatol. 2020;18:2226-2233.e4. [PubMed] [DOI] [Cited in This Article: ] |
| 56. | Cohen S, Booth GH Jr. Gastric acid secretion and lower-esophageal-sphincter pressure in response to coffee and caffeine. N Engl J Med. 1975;293:897-899. [PubMed] [DOI] [Cited in This Article: ] |
| 57. | Kim J, Oh SW, Myung SK, Kwon H, Lee C, Yun JM, Lee HK; Korean Meta-analysis (KORMA) Study Group. Association between coffee intake and gastroesophageal reflux disease: a meta-analysis. Dis Esophagus. 2014;27:311-317. [PubMed] [DOI] [Cited in This Article: ] |
| 58. | Brenner H, Rothenbacher D, Bode G, Adler G. Relation of smoking and alcohol and coffee consumption to active Helicobacter pylori infection: cross sectional study. BMJ. 1997;315:1489-1492. [PubMed] [DOI] [Cited in This Article: ] |
| 59. | Zhu Q, Hua L, Chen L, Mu T, Dong D, Xu J, Shen C. Causal association between obstructive sleep apnea and gastroesophageal reflux disease: A bidirectional two-sample Mendelian randomization study. Front Genet. 2023;14:1111144. [PubMed] [DOI] [Cited in This Article: ] |
| 60. | Li L, Ren Q, Zheng Q, Bai Y, He S, Zhang Y, Ma H. Causal associations between gastroesophageal reflux disease and lung cancer risk: A Mendelian randomization study. Cancer Med. 2023;12:7552-7559. [PubMed] [DOI] [Cited in This Article: ] |
| 61. | Adewuyi EO, O'Brien EK, Porter T, Laws SM. Relationship of Cognition and Alzheimer's Disease with Gastrointestinal Tract Disorders: A Large-Scale Genetic Overlap and Mendelian Randomisation Analysis. Int J Mol Sci. 2022;23. [PubMed] [DOI] [Cited in This Article: ] |