Published online May 27, 2016. doi: 10.5496/wjmg.v6.i2.17
Peer-review started: April 19, 2016
First decision: May 10, 2016
Revised: May 15, 2016
Accepted: May 18, 2016
Article in press: May 20, 2016
Published online: May 27, 2016
We report a 28-year-old female who presented with severe joint pain, chronic muscle weakness, Raynaud’s phenomenon, and hypermobility. She was found to have a 6074A > T nucleotide transition in the TNXB gene causing an amino acid protein change at Asp2025Val classified as likely pathogenic. We add this clinical report to the literature and classical human disease gene catalogs to identify this specific mutation as disease-causing. This gene variant was reported previously in a different 36-year-old patient who shared our patient’s symptoms of joint hypermobility, skeletal and joint pain, skin elasticity and musculoskeletal problems, thereby causing a more severe presentation than seen in the hypermobility type of Ehlers-Danlos syndrome (EDS). At the time of writing, a few mutations in the TNXB gene have been recognized as pathogenic causing EDS due to tenascin-X deficiency, but the variant identified in our patient has not been recognized as pathogenic in online genetic databases. Our case study in combination with peer-reviewed literature suggests that the 6074A > T nucleotide transition in the TNXB gene may be classified as disease-causing for EDS due to tenascin-X deficiency.
Core tip: Various types of Ehlers-Danlos syndrome (EDS) have unique phenotypic features and genetic causes that are under investigation. This case report presents a gene variant (6074A > T nucleotide transition in the TNXB gene) not previously classified as disease-causing which we propose should be classified as pathogenic. This variant appears to produce joint hypermobility, skeletal pain, and musculoskeletal problems and should be classified as causing EDS due to tenascin-X deficiency.
- Citation: Kaufman CS, Butler MG. Mutation in TNXB gene causes moderate to severe Ehlers-Danlos syndrome. World J Med Genet 2016; 6(2): 17-21
- URL: https://www.wjgnet.com/2220-3184/full/v6/i2/17.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v6.i2.17
Ehlers-Danlos syndrome (EDS) encompasses a heterogeneous collection of connective tissue disorders with joint, skin and vascular involvement. Patients diagnosed with EDS have diverse clinical findings, which may be better understood and classified by identifying the genetic contribution to symptoms[1,2]. Although this condition has been recognized since 1901[3] and is now classified into nine types, delineation of the types and relationships to causative genes and their variants are underway, impacted by advances in genetic technology and next generation (exome) sequencing[4,5]. Here, we assert that mutations in the TNXB gene, which encodes the glycoprotein tenascin-XB and related to the benign hypermobility form of EDS, may cause a more severe form or presentation of EDS.
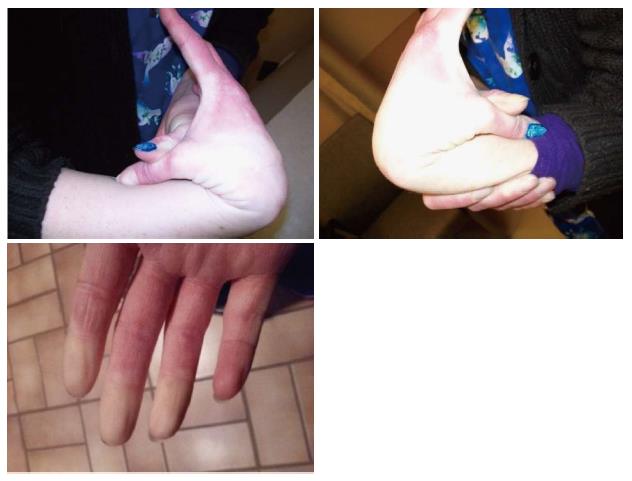
Our 28-year-old patient showed no developmental delays as a child. She did undergo a right knee arthroscopic surgery not related to an injury at 18 years of age and breast augmentation at 21 years of age. She has had two pregnancies and was anemic during both pregnancies, due to decreased red blood cell count. The pregnancies were full-term and labor and delivery were normal. She experienced increased joint hypermobility during both of her pregnancies. Previous lab work showed normal thyroid and hepatic function. The patient had generalized joint pain since childhood with mild scoliosis. Anti-inflammatory agents did not relieve her pain. She has experienced instability of the right shoulder, pelvis and both hip joints with more problems with advancing age. During the clinic visit, we completed an assessment of the patient’s joint mobility using the Beighton scale[6], which results in a numerical score from 0 to 9 with one point assigned for the ability to perform each of the following actions: Passive dorsiflexion of the little finger beyond 90 degrees; passive apposition of the thumb to the flexor aspects of the forearm; hyperextension of the elbow beyond 10 degrees; hyperextension of the knee beyond 10 degrees; and forward flexion of the trunk, with knees straight, so that palms of hands rest easily on the floor. Our patient’s Beighton score was 6 out of 9, including easy placement of palms to the floor (one point), hyperextension of both knees (two points), hyperextension of one elbow (one point), and passive apposition of both thumbs to the forearms (two points) as shown in Figure 1.
Furthermore, our patient also experienced Raynaud’s phenomenon, manifested as paresthesia with decreased blood flow in her fingers, lip, ears, and nose (Figure 1). This clinical presentation led to rheumatology evaluation including antibody studies and SED rate to diagnose a mixed connective tissue disorder such as lupus or rheumatoid arthritis. The results were normal. The
patient’s back pain continues to worsen and she experiences weakness throughout much of her body. She is unable to participate in mildly strenuous manual labor or movement related activities. Our patient identifies chronic pain and generalized muscle weakness as the most significant medical problems detrimental to daily functioning and quality of life.
The family history was limited due to little to no contact with her siblings and parents, all of whom live in Israel. Our patient has 100% Ashkenazi Jewish ancestry but has not had any Jewish specific genetic disease testing. She has a son who is 4 years of age and a daughter who is 10 years of age, both currently without relevant medical concerns or health problems.
We ordered next generation DNA sequencing panel and deletion/duplication analysis of 33 genes related to connective tissue disorders (ABCC6, ACTA2, ATP7A, CBS, CHST14, COL1A1, COL1A2, COL2A1, COL3A1, COL5A1, COL5A2, COL9A1, COL9A2, COL9A3, COL11A1, COL11A2, ELN, FBN1, FBN2, FKBP14, FLNA, MYH11, MYLK, PLOD1, SKI, SLC2A10, SLC39A13, SMAD3, SMAD4, TGFB2, TGFBR1, TGFBR2, and TNXB) performed commercially at the University of Nebraska Medical Center (Omaha, NE) using a blood sample from our patient. The only gene variant identified was a 6074A > T transition in the TNXB gene causing a Asp2025Val amino acid change at the protein level, classified as a variant of uncertain clinical significance. No known pathogenic deletions or duplications were detected within these genes.
Classification for EDS began in the late 1960s[4] and continues to evolve and expand over time, as diagnoses move beyond focusing solely on clinical symptoms to a more genomics-driven approach. A popular simplified classification based on the Villenfranche nosology outlined the following six major types of EDS[4]: Classical (Type Iand Type II) with defect in type 5 collagen; Hypermobile (Type III) with unknown genetic causation; Vascular (Type IV) with abnormal type 3 collagen; Kyphoscoliosis (Type VI) with deficiency of lysyl hydroxylase (LH); Arthrochalasia (Type VII) with deficiency of chains in type I collagen; and Dermatosparaxis (Type VII) with deficiency of procollagen N-proteinase enzyme in type I collagen.
In a more recent review[5], this classification was expanded based on current evidence, and a total of nine types of EDS were outlined: Classical (caused by defects in collagen COL5A1 and COL5A2 genes) involving skin laxity, joint hypermobility, widened scars, and easy bruising; Hypermobile (unknown cause) with velvet skin and laxity, joint dislocations and hypermobility; Vascular (COL3A1 gene) with arterial rupture, hypermobility, varicose veins, thin translucent skin, and easy bruising; Kyphoscoliosis (LH 1 - PLOD1 gene) with muscle hypotonia in infants, scoliosis from birth, joint laxity, gross motor delay, easy bruising, and fragility of sclerae; Arthrochalasia (collagen 1 - COL1A1, COL1A2 genes) with severe joint hypermobility, congenital bilateral hip dislocations, skin elasticity, easy bruising, kyphoscoliosis, and osteopenia; Dermatosparaxis (procollagen N-proteinase - ADAMTS2 gene) with sagging redundant skin, blue sclerae, short stature, and umbilical hernia; EDS with scoliosis, myopathy, hearing impairment (FKBP14 gene); Musculocontractural EDS (CHST14 gene) with progressive kyphoscioliosis, adducted thumbs in infancy, joint hypermobility, clubfoot, arachnodactyly, elastic skin, and poor wound healing; and finally Tenascin-X deficient (TNXB gene). Those with Tenascin-X (TNXB) deficient EDS involve features of marked skin laxity, pronounced joint hypermobility and severe bruising[5]. We extend the description of those with mutations in the TNXB gene to include significant joint pain and Raynaud’s phenomenon, thus resulting in a more severe form of EDS than previously classified. Whether the severity depends on the type or location of the gene variant (mutation) is unknown.
The TNXB gene (sometimes known by other names such as HXBL, TNX, TENX, XB) belongs to a family of genes called fibronectin type III domain containing and localizes to the major histocompatibility complex (MHC) class III region on chromosome 6, at cytogenetic location 6p21.3. The TNXB gene encodes for a protein called tenascin-X, which may regulate assembly and production of specific types of collagen. Tenascin-X helps organize and maintain the structure of tissues supporting the joints, organs, skin, connective tissues, and muscles[7].
Tenascin-X is an extracellular matrix glycoprotein, thought to function in matrix maturation during wound healing[7] and became the first EDS gene that does not encode a collagen-modifying enzyme or a fibrillar collagen[8]. Mice knockouts (Tnxb-/-) showed skin hyperextensibility, reflecting the human phenotype of EDS, and the skin collagen content was significantly reduced compared to wildtype. Additionally, the density of fibrils in the skin of knockout (Tnxb-/-) mice was reduced compared to wildtype, suggesting tenascin-X plays a significant regulatory role in collagen deposition by dermal fibroblasts[8]. Hence, tenascin-X appears to be involved in the regulation of the structure and stability of elastic fibers, which give elasticity and flexibility to connective tissues.
Alterations in the TNXB gene and effects on protein levels have also been studied[9]. The researchers measured tenascin-X in serum from 21 healthy subjects and 151 patients diagnosed with EDS. They reported that 146 of the 151 patients with EDS had detectable tenascin-X levels in their serum. A closer examination of the 5 patients diagnosed with EDS and lacking serum tenascin-X revealed that all had hyperelastic skin and hypermobility, and a subset had joint pain and multiple subluxations. One patient was homozygous and another heterozygous for a 30-kb deletion in the TNXB gene resulting in a nonfunctional fusion gene. Two patients were homozygous for a 2-bp deletion in exon 8 of the TNXB gene, which altered the open reading frame coding for amino acids 1184 through 1230 before causing a premature stop codon. The fifth patient was homozygous for a 2-bp insertion in exon 3 which caused a stop codon at position 707. These genetic variations are now classified as causing EDS specifically due to tenascin-X protein deficiency.
Furthermore, a different study[10] identified three missense mutations in the TNXB gene, one in each of three patients diagnosed with the hypermobility type EDS but with normal tenascin-X serum levels: A 3582A > G transition (Val1195Met), an 85C > T transition (Arg29Trp), and a 12097C > A transition (Leu4033Ile). These wild type amino acids and locations are strongly conserved in this protein. The researchers then compared skin biopsies from these patients with other EDS hypermobility type patients who did not have gene variants in the TNXB gene and determined that elastic fiber abnormalities in hypermobility type EDS are specific for TNX-haploinsufficient individuals and confirm an important role for TNX in regulating elastic fiber integrity in humans[10].
As demonstrated in the literature by examples described, a variety of mutations, deletions, and insertions in the TNXB gene may cause features of EDS. Our patient’s gene change was classified as a variant of unknown significance and not as yet classified as disease-causing. This missense mutation in exon 17 of the TNXB gene in our patient is moderately conserved with an in silico prediction of trending damaging. This mutation causes a significant amino acid change from an aspartic acid which is negatively charged to a valine which is non-polar; this transition could theoretically impact protein structure due to such a dramatic change in the amino acid classification type. Based on our patient’s clinical symptoms and DNA findings with evidence from the literature, we propose that the 6074A > T nucleotide transition (p.Asp2025Val) identified in our patient causes a moderate to severe form of EDS that includes significant joint pain, muscle weakness, hypermobility and Raynaud’s phenomenon.
A limitation of our report is the lack of contact by the patient with her siblings or parents, all of whom live in Israel. Therefore, it is not possible for us to reach members of her family to identify the presence or absence of the gene variant and to better characterize the genotype-phenotype relationship. Furthermore, our patient lives at a considerable distance from the medical center making travel difficult for her and her immediate family (e.g., younger children). It is unlikely that arrangements could be made in a timely fashion to undertake skin biopsy and assessment of elastic fibers in the patient or her living children without evidence of disease in which an invasive procedure is not warranted even if accessible.
Our patient is also of Ashkenazi Jewish ancestry. A 36-year-old patient with this same amino acid change at the protein level reported to be likely pathogenic[2,11-14] and that patient was diagnosed clinically with the hypermobility type of EDS. The patient’s symptoms included hypermobility, skeletal pain, skin elasticity, gastrointestinal and musculoskeletal problems, all consistent with EDS. The status of Jewish ancestry with this reported patient is unknown. We propose that both of these patients may have a form of EDS due to their specific mutation in the TNXB gene, causing more severe symptoms sufficiently significant to interfere with daily functioning and quality of life. More research is needed to conclude that this gene variant is causative and authors encourage the reporting of other patients with variants of the TNXB gene, clinical features and genotype-phenotype correlations.
We thank our patient.
A 28-year-old female presented with joint pain, muscle weakness, hypermobility, and Raynaud’s phenomenon. She was referred to our clinic for evaluation for a connective tissue disorder.
Severe joint pain, chronic muscle weakness, Raynaud’s phenomenon, hypermobility with Beighton hypermobility score of 6 out of 9, mild scoliosis.
Ehlers-Danlos syndrome.
Previous rheumatology evaluation ruled out lupus or rheumatoid arthritis through antibody studies and SED rate. Testing through the University of Nebraska Medical Center (Omaha, NE) showed a nucleotide 6074A > T transition in the TNXB gene using a blood sample.
Reduce daily stress on joints by minimizing mildly strenuous activity. Avoid hyper-extending joints.
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, Fox M, Fogel BL, Martinez-Agosto JA, Wong DA, Chang VY, Shieh PB, Palmer CG, Dipple KM, Grody WW, Vilain E, Nelson SF. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014; 312: 1880-1887 [PMID: 25326637 DOI: 10.1001/jama.2014.14604].
Ehlers-Danlos syndrome includes a heterogeneous collection of connective tissue disorders with vascular, skeletal and skin involvement.
As technology advances, authors are able to order more sophisticated genetic testing in the clinical setting, such as the sequencing and deletion/duplication panel ordered in this case report. However, the meaning of the results is not always clear, as more often than not they identify “variants of unknown clinical significance”, meaning the change at the DNA level does not have a clear, recognized outcome for the phenotype. In these cases, they use computer simulation to predict whether or not the change may be harmful to protein structure or function. They also review literature for cases involving the variant, with the hope of associating genotype with phenotype. In the case of the patient reported in this manuscript, they concluded through in-depth examination of the variant and a comprehensive literature review that the 6074A > T transition in the TNXB gene seen in the patient is likely pathogenic and causes a moderate to severe form of Ehlers-Danlos syndrome.
The authors report a case of Ehlers-Danlos syndrome with a mutation in TNXB gene. The report is interesting.
P- Reviewer: Castori M, Sakamoto A, Sakkas LI S- Editor: Qiu S L- Editor: A E- Editor: Lu YJ
| 1. | Online Mendelian Inheritance in Man Johns Hopkins University, Baltimore, MD. [updated 2014 Aug 26]. Available from: http//omim.org/entry/606408?search=tnxb&highlight=tnxb. [Cited in This Article: ] |
| 2. | Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880-1887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 670] [Cited by in F6Publishing: 704] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 3. | Ehlers E. Cutis laxa, neigung zu harmorrhagien in der haut, lockerun mehrer Artikulationen. Dermat Ztschr. 1901;8:173. [Cited in This Article: ] |
| 4. | Parapia LA, Jackson C. Ehlers-Danlos syndrome--a historical review. Br J Haematol. 2008;141:32-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 5. | Sobey G. Ehlers-Danlos syndrome: how to diagnose and when to perform genetic tests. Arch Dis Child. 2015;100:57-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 6. | Beighton P, Solomon L, Soskolne CL. Articular mobility in an African population. Ann Rheum Dis. 1973;32:413-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1068] [Cited by in F6Publishing: 991] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 7. | National Library of Medicine (US). Genetics Home Reference [Internet]. Bethesda (MD): The Library; 2016; Mar 28. TNXB. [reviewed 2015 Nov; cited 2016 Apr 7] Available from: http//ghr.nlm.nih.gov/gene/TNXB. [Cited in This Article: ] |
| 8. | Mao JR, Taylor G, Dean WB, Wagner DR, Afzal V, Lotz JC, Rubin EM, Bristow J. Tenascin-X deficiency mimics Ehlers-Danlos syndrome in mice through alteration of collagen deposition. Nat Genet. 2002;30:421-425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 192] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 9. | Schalkwijk J, Zweers MC, Steijlen PM, Dean WB, Taylor G, van Vlijmen IM, van Haren B, Miller WL, Bristow J. A recessive form of the Ehlers-Danlos syndrome caused by tenascin-X deficiency. N Engl J Med. 2001;345:1167-1175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 306] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 10. | Zweers MC, Dean WB, van Kuppevelt TH, Bristow J, Schalkwijk J. Elastic fiber abnormalities in hypermobility type Ehlers-Danlos syndrome patients with tenascin-X mutations. Clin Genet. 2005;67:330-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Beighton P, Horan F. Orthopaedic aspects of the Ehlers-Danlos syndrome. J Bone Joint Surg Br. 1969;51:444-453. [PubMed] [Cited in This Article: ] |
| 12. | Beighton P, De Paepe A, Steinmann B, Tsipouras P, Wenstrup RJ. Ehlers-Danlos syndromes: revised nosology, Villefranche, 1997. Ehlers-Danlos National Foundation (USA) and Ehlers-Danlos Support Group (UK). Am J Med Genet. 1998;77:31-37. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | De Paepe A, Malfait F. The Ehlers-Danlos syndrome, a disorder with many faces. Clin Genet. 2012;82:1-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 242] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 14. | Juul-Kristensen B, Røgind H, Jensen DV, Remvig L. Inter-examiner reproducibility of tests and criteria for generalized joint hypermobility and benign joint hypermobility syndrome. Rheumatology (Oxford). 2007;46:1835-1841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 181] [Article Influence: 10.6] [Reference Citation Analysis (0)] |