Published online Apr 24, 2024. doi: 10.5306/wjco.v15.i4.531
Peer-review started: December 27, 2023
First decision: January 15, 2024
Revised: January 29, 2024
Accepted: March 7, 2024
Article in press: March 7, 2024
Published online: April 24, 2024
Metastasis remains a major challenge in the successful management of malignant diseases. The liver is a major site of metastatic disease and a leading cause of death from gastrointestinal malignancies such as colon, stomach, and pancreatic cancers, as well as melanoma, breast cancer, and sarcoma. As an important factor that influences the development of metastatic liver cancer, alternative splicing drives the diversity of RNA transcripts and protein subtypes, which may provide potential to broaden the target space. In particular, the dysfunction of splicing factors and abnormal expression of splicing variants are associated with the occurrence, progression, aggressiveness, and drug resistance of cancers caused by the selective splicing of specific genes. This review is the first to provide a detailed summary of the normal splicing process and alterations that occur during meta
Core Tip: Metastatic liver cancer refers to tumors formed outside the liver that metastasize to the liver and colonize it. Abnormal alternative splicing is a molecular characteristic unique to almost all tumor types. Most tumors exhibit a wide range of splicing abnormalities compared to the surrounding healthy tissues. This review is the first to provide a detailed summary of the normal splicing process and alterations that occur during metastatic liver cancer by examining splicing factor changes, abnormal splicing, and the contribution of hypoxia to cellular changes.
- Citation: Geng DY, Chen QS, Chen WX, Zhou LS, Han XS, Xie QH, Guo GH, Chen XF, Chen JS, Zhong XP. Molecular targets and mechanisms of different aberrant alternative splicing in metastatic liver cancer. World J Clin Oncol 2024; 15(4): 531-539
- URL: https://www.wjgnet.com/2218-4333/full/v15/i4/531.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i4.531
Primary liver cancer, also known as hepatocellular carcinoma (HCC), originates in the liver and is often associated with chronic liver diseases such as cirrhosis, infections with hepatitis B or C viruses, or alcohol-related liver disease[1]. Conversely, metastatic liver cancer refers to tumors formed outside the liver that metastasize to the liver and colonize it. Owing to the dual blood supply from the hepatic artery and portal vein, the liver has become the most common parenchymal organ to which most malignant tumors metastasize[2]. Metastatic cancer has become a major clinical challenge because of its high incidence and poor prognosis. Metastatic liver cancer, or liver metastasis, is caused by the spread of cancer cells from other primary sites (such as the colon, rectum, stomach, and breast) to the live[3-6]. The prognosis of patients with liver metastasis varies depending on the type of primary cancer. Liver metastasis in some cancers, such as lung cancer, is associated with a poor prognosis[7].
Currently, the treatment of metastatic liver cancer is completely different from that of the primary cancer. Although tumors grow in the liver, the biological activity of metastatic liver cancer is different from that of tumors at the primary site, and liver metastasis has the characteristics of multifocal and late-stage diseases[7]. The treatment of metastatic liver cancer usually involves systemic treatments such as chemotherapy and targeted therapy[3-6]. Therefore, initially determining the organ or tissue source of the primary cancer is necessary (to obtain pathology findings) and then use systemic treatment (choosing a plan based on the pathology of the primary cancer) and local liver resection, including surgical, ablation, and systemic treatment methods[7-10]. The combination of minimally invasive image-guided therapies, such as radioembolization and percutaneous liver-guided therapy, has expanded the treatment options for patients with obvious metastatic liver disease. However, further research is required to optimize the timing and safety of combining systemic and local regional therapies. Metastatic liver cancer presents a complex clinical environment with different primary cancer origins, prognostic impacts, and challenges in accurate diagnosis and management. Understanding the metastatic patterns, prognostic factors, and immune microenvironments of liver metastases is crucial for developing effective treatment strategies and improving patient prognosis[11-13].
Abnormal alternative splicing (AS) is a molecular characteristic unique to almost all tumor types[14]. Most tumors exhibit a wide range of splicing abnormalities compared to the surrounding healthy tissues, including frequent retention of normally excised introns, inappropriate expression of isoforms that are typically limited to other cell types or developmental stages, splicing errors that damage tumor suppressor genes or promote oncogenic gene expression, and promotion of tumor development through various mechanisms, including increased cell proliferation, reduced apoptosis, enhanced migration and metastasis, drug-resistant chemotherapy, and evasion of immune monitoring[15,16]. Metastatic liver cancer undergoes significant changes over time. In cancer cells derived from the liver and bile ducts, abnormal proteins are synthesized due to abnormal splicing associated with cancer. This leads to the dysproliferation of these cells, ultimately transforming them into invasive, migratory, and multidrug-resistant phenotypes, resulting in a poor prognosis for these liver cancers[8,17,18].
In this review, we highlight the recent developments in AS events. We will also describe the regulation of AS in primary and metastatic liver cancers. In addition, this review integrates the biological functions of AS and splicing products as well as current efforts to develop their potential for clinical application in the diagnosis or treatment of cancer.
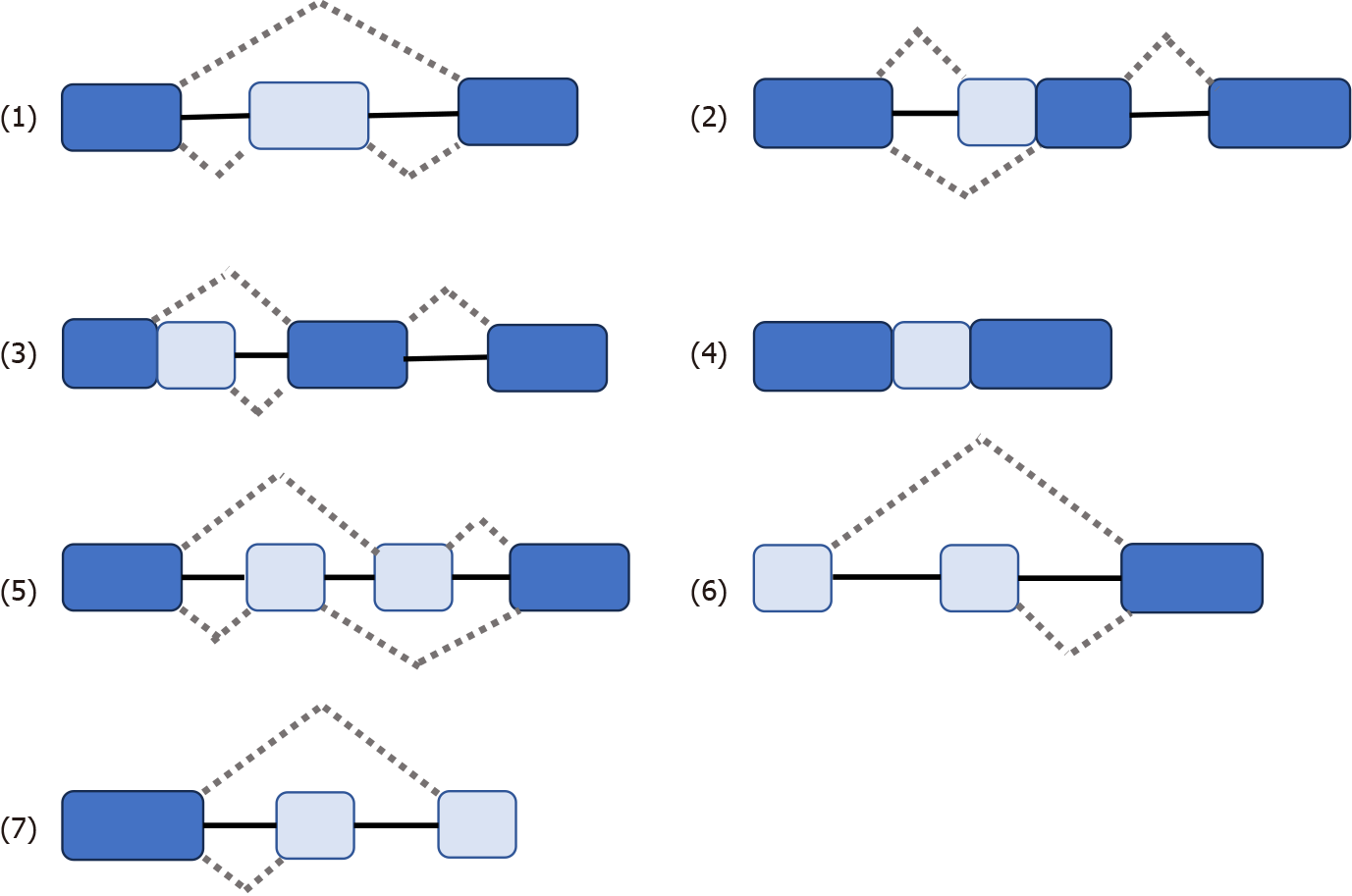
Some genes have one mRNA precursor that produces different mRNA splicing isomers using different splicing methods (choosing different splicing sites) in a process known as variable splicing (or AS)[14,15,18-20]. Variable splicing is the most common and widespread type of splicing[14]. Variable splicing is an important mechanism for regulating gene expression and generating proteomic diversity and is an important reason for the large differences in the number of genes and proteins in eukaryotes[21]. In vivo, there are seven types of variable splicing: (1) Exon skip; (2) Retained intron; (3) Alternate Donor site; (4) Alternate acceptor site; (5) Alternate promoter; (6) Alternate terminator; and (7) Mutually exclusive exons[14,15,17] (Figure 1). During variable splicing, the different exons of a gene sequence are selectively linked to form multiple transcripts. Consequently, the same gene can encode many different proteins, thereby increasing the functional diversity of the gene. This type of splicing is very common in mammals and is found in more than 90% of genes[22]. The development of metastatic cancer is influenced by multifactorial conditions, possibly due to: (1) Altered expression of the spliceosome; (2) mutations affecting genes encoding spliceosome components and related regulatory proteins; (3) disruption of splice site or splicing regulatory sites (enhancers or silencers); and (4) impaired signaling pathways involved in the regulation of splicing mechanisms[17,23,24].
Primary HCC tumor tissue exhibits a high degree of differential splicing compared to normal liver tissue. A growing body of research has shown that alterations in the splicing program in HCC tumor cells generate novel protein subtypes that often have different and sometimes opposite functions to their classical counterparts[25]. These changes were significantly associated with patient survival[12]. These findings suggest that AS plays a crucial role in HCC progression and prognosis. Primary liver cancer includes various tumors such as HCC and intrahepatic cholangiocarcinoma (iCCA)[26]. One study showed that differences between HCC and iCCA AS affected hundreds of genes[19]. Thus, alternative and tumor-specific subtypes caused by abnormal splicing are common during liver tumorigenesis[21,27].
AS disorder is also associated with the pathogenesis of liver cancer. For example: Loss of SRSF3 induces IGF2 expression and altering INSR splicing to allow insulin-like growth factor II (IGF2) signaling to be conducted through insulin receptor (IR)-A in hepatocytes[28]. Hepatic IGF2 expression is a carcinogenic driver in aging-related HCC mouse models, causing DNA damage and supporting hepatocyte proliferation. This allowed for the accumulation of somatic mutations. EGFR regulates the selective splicing of IR pre-mRNA in HCC cells. After ligand binding, EGFR activation triggers an intracellular signaling cascade, which implies MEK activation. This stimulates the transcription of genes encoding different splicing factors, namely CUGBP1, hnRNPH, hnRNPA1, hnRNPA2B1, and SF2/ASF. hnRNPF expression is not regulated by the EGFR-dependent pathway. Interaction between splicing factors and IR pre-mRNA promotes the selective splicing of IR exon 11. Consequently, the expression of the IR-A subtype increases to the detriment of IR-B, which allows for the transmission of proliferative signals in response to insulin and IGF2, leading to HCC development[29].
In summary, the dysregulation of AS in liver cancer has been shown to affect various molecular pathways, underscoring the influence of AS dysregulation on the molecular mechanisms of liver cancer development and its extensive involvement in the pathogenesis, progression, and prognosis of HCC. The replacement of gene products produced by abnormal splicing has been linked to positive effects in cancer, making AS a potential target for gene therapy[30]. These findings suggest that understanding AS in liver cancer may lead to the development of novel therapeutic interventions.
Pathogenesis of metastatic liver cancer involves a complex interplay of molecular mechanisms, including the role of splicing factors in cancer progression, AS, and hypoxia-induced splicing changes[31].
The role of splicing factors in metastatic liver cancer has attracted increasing interest in cancer research. Splicing factors, including those in metastatic liver cancer, play a direct role in cancer development[32]. Abnormal RNA splicing has been recognized as a driver of cancer development and changes in AS of RNA have been associated with liver cancer progression[26]. In addition, alterations in splicing are associated with liver cancer markers, including de-differentiation and genomic instability, which are the core processes of tumor transformation[16].
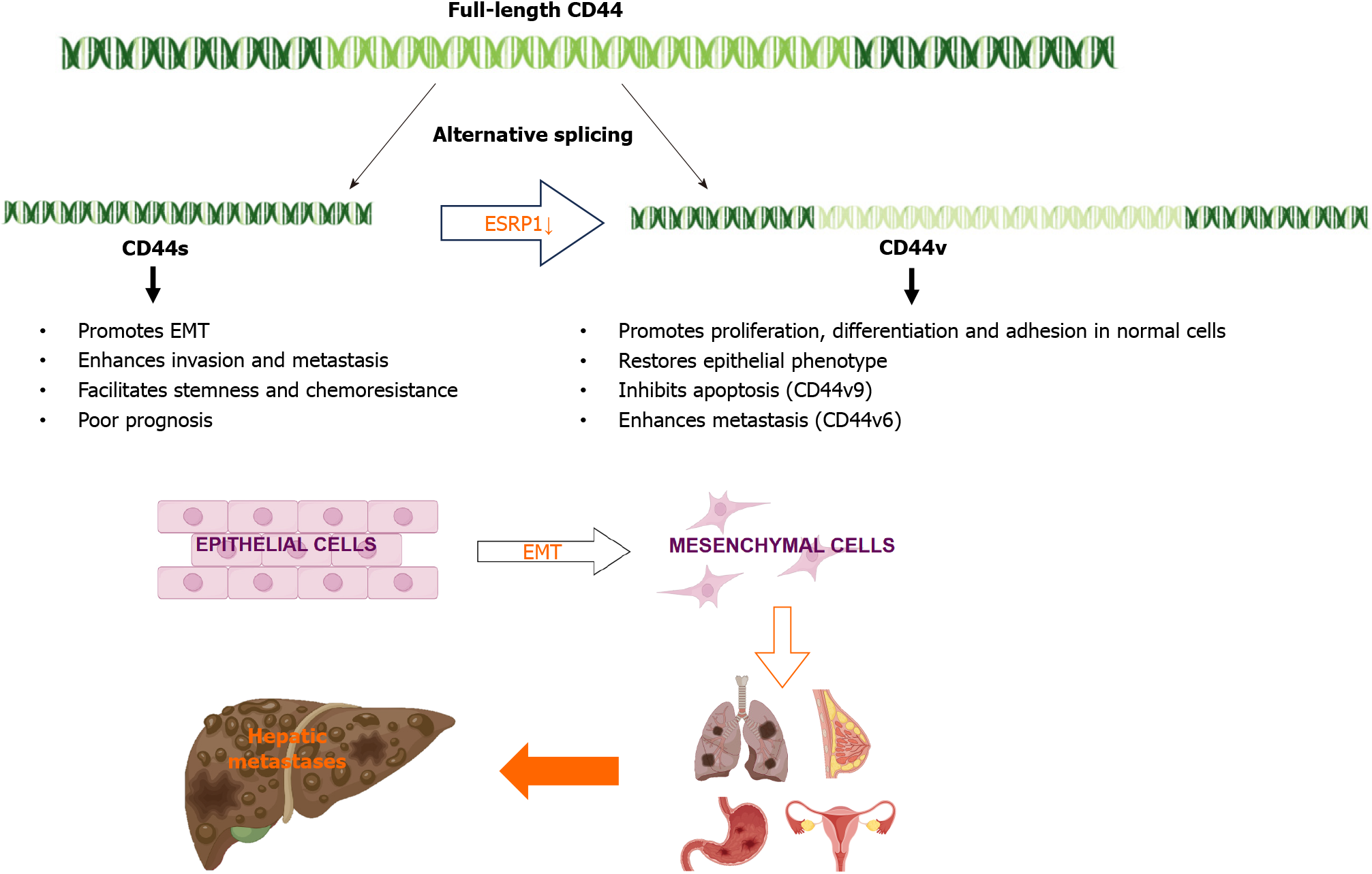
In liver cancer, the AS of specific genes has been shown to contribute to cancer progression and metastasis. For example, epithelial splicing regulatory protein 1 (ESRP1) plays a key role in the regulation of CD44 AS[33]. ESRP1 is an epithelium-specific splicing factor that regulates the AS of several genes, including fibroblast growth factor receptor 2 and CD44[34]. Epithelial-mesenchymal transition (EMT) is a specific biological process in which epithelial cells are transformed into stromal cells. It is important for epithelial cell-derived malignant tumor cells to acquire migration and invasion abilities. EMT plays a crucial part in embryonic development, chronic inflammation, tissue reconstruction, cancer metastasis, and various fibrotic diseases[35]. The main characteristics of EMT include reduced expression of cell adhesion molecules (such as E-cadherin), transformation of the cytoskeleton from keratin to vimentin, and altered morphological characteristics of mesenchymal cells. Through EMT, epithelial cells lose cell polarity, their connection to the basement membrane, and other epithelial phenotypes, while gaining higher interstitial phenotypes, such as migration and invasion, apoptosis inhibition, and degradation of the extracellular matrix[36]. During EMT, ESRP1 expression is sharply reduced, facilitating the transition from variant CD44 (CD44v) to standard CD44 (CD44s), mediating the expression of isoforms required for EMT. ESRP1 promotes liver metastasis in breast cancer cells by enhancing EMT[34]. ESRP1 regulates subtype conversion and determines gastric cancer metastasis[37]. ESRP1 drives AS of CD44, thereby enhancing invasion and migration of epithelial ovarian cancer cells[38]. ESRP1 has been identified as a favorable prognostic factor for pancreatic cancer[39], alleviating pancreatic metastasis. In contrast, the silencing of ESRP1 has been shown to drive the malignant transformation of human lung epithelial cells[40], suggesting that cancer progression is strongly influenced by splicing factors (Figure 2).
AS events have been identified as prognostic factors for HCC, highlighting their potential impact on the development and prognosis of liver cancer[41]. Various AS events may also influence the development of metastatic liver cancer. Studies have shown that abnormal AS events promote malignant cancer progression[42].
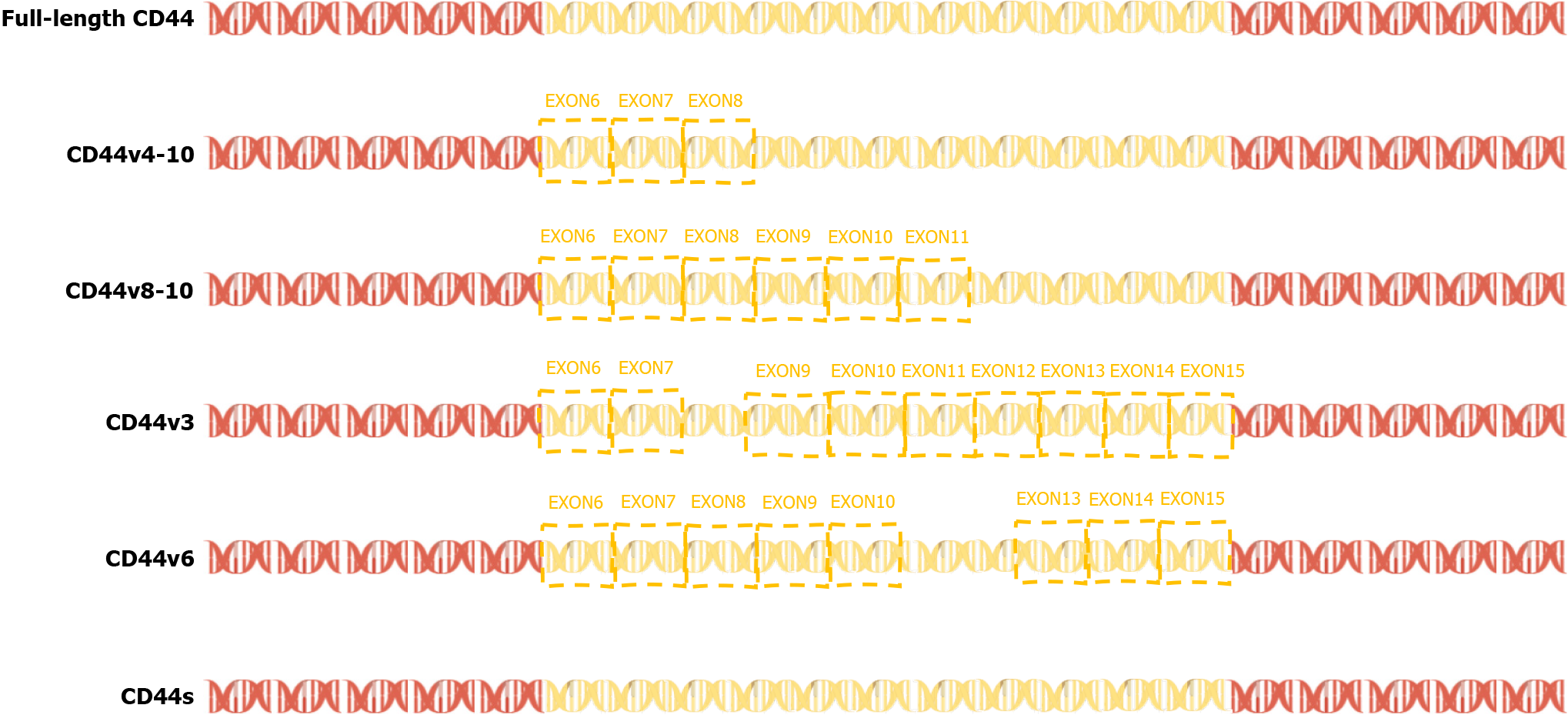
In 2015, a team found that PC-3 and its derived cell lines crossed the transfer barrier in vitro and in vivo, providing an excellent, unbiased system for comprehensively characterizing AS events and identifying the key splicing factors that influence the splicing regulation of transfer. This suggests that partially selective splicing events are associated with metastatic colonization of cancer cells, suggesting a potential role in promoting metastasis[43]. Liver metastasis may occur in different tumors, and different splicing events may promote liver metastasis. For example, the splicing mediated by RBFOX2 shifts from an epithelial-specific event to a mesenchymal-specific event, leading to a higher degree of tissue invasion, which in turn leads to liver metastasis[44]. Different splicing subtypes of the same spliceosome, such as CD44, promote liver metastasis. CD44 is a cell surface glycoprotein involved in cancer progression and metastasis. AS of CD44 mRNA produces various subtypes, including CD44s and CD44v (Figure 3), which are associated with cancer metastasis. The CD44-ZEB1-ESRP1 feedback loop can control the cell phenotype and prognosis of patients with cancer by determining the CD44 subtype expression[45]. Some splicing events that lead to liver cancer metastasis, such as the overexpression of IGF2 and decreased splicing activity of SRSF3, are considered major causes of DNA damage and drivers of liver cancer, indicating the importance of specific splicing factors in liver cancer development. While affecting the process of liver metastasis, it also affects changes in tumor drug resistance. The FUS/circEZH2/KLF5 feedback loop promotes liver metastasis of breast cancer by FUS promoting the reverse splicing process of circEZH2 by binding to the 3’-lateral intron portion of pre-EZH2 to enhance the EMT, and may also influence drug resistance of liver metastases through this mechanism[46]. In summary, AS events are involved in the occurrence and development of metastatic liver cancer, highlighting the importance of splicing regulation in cancer progression and metastasis.
Hypoxia is associated with changes in EMT, angiogenesis, local tissue invasion, endothelium, exocytosis, and pre-metastatic niche formation[47]. Hypoxia, a hallmark of the tumor microenvironment, induces AS, thereby promoting the invasive behavior of cancer cells[31]. Hypoxia inhibits cancer cell differentiation and promotes cancer cell invasion and metastasis, emphasizing its role in promoting cancer cell metastasis[48]. Hypoxia-induced splicing changes play a crucial role in the occurrence and progression of cancer metastasis. Hypoxia-induced selective splicing is cell type-specific and has highly conserved universal target genes, indicating that hypoxia has a broad impact on splicing[49]. In the DNA damage response, hypoxia drives the selective splicing of genes towards non-coding subtypes by increasing intron retention[50]. Similarly, hypoxia promotes the expression of splicing subtypes of Myc-related factor X in endothelial cells, mediated by nonsense decay degradation, and another splicing subtype that encodes unstable proteins[51].
Hypoxia leads to significant changes in the selective splicing of prostate cancer cells and increased expression of CLK splicing factor kinase, leading to liver metastasis[52]. In addition, hypoxia regulates CD44 and its variant subtypes through HIF-1α in triple-negative breast cancer, highlighting the role of hypoxia in regulating various splicing events associated with cancer progression[48].
The effect of hypoxia on AS has been recognized as a powerful driving force for tumor pathogenesis and progression, and various studies have emphasized the important influence of hypoxia-induced splicing changes on the pathogenesis of metastatic liver cancer. Understanding the molecular mechanisms underlying hypoxia-induced splicing changes is essential for developing targeted therapeutic strategies to mitigate the invasive behavior of metastatic liver cancer cells and improve patient outcomes.
The liver has a rich blood supply, so it provides fertile “soil” for metastasis to spread[53]. The liver is one of the largest blood vessel networks in the body. It receives blood from the gut, which contains a lot of nutrients. The blood vessels at the end of the liver also have high pressure, so it is easier to accommodate and colonize metastasized cancer cells[54]. The most common source of metastatic liver cancer is colorectal cancer, followed by pancreatic, breast, melanoma and lung cancer. The common ways of metastasis include direct invasion, lymphatic metastasis and blood-derived metastasis. Malignant tumors that directly invade the organs and tissues around the liver, such as gastric cancer, gallbladder cancer, pancreatic cancer, colon cancer and duodenal cancer. Lymphatic metastasis is more common in digestive system malignancies, pelvic or retroperitoneal malignancies, breast cancer, lung cancer and gallbladder cancer. Hematogenous metastasis can also be further subdivided into hepatic artery and portal vein metastasis. Any tumor cells entering the liver through these vessels can cause liver metastasis, such as esophageal and gastrointestinal tumors and some sarcomatoid tumors with higher malignant degree.
Metastatic liver cancer presents significant clinical challenges owing to its aggressiveness, poor prognosis, and limited treatment options. Studies based on the SEER database emphasize that patients with primary extrahepatic metastases have poor prognosis[55]. In 2020, a practical study of high-intensity focused ultrasound ablation in 250 patients, including a primary liver cancer cohort (n = 80) and metastatic liver cancer cohort (n = 195), yielded 1-year survival rates of 70.69% and 48.00%, respectively[56]. These findings highlight the need for innovative therapeutic modalities to address the adverse effects of metastatic liver cancer. Metastatic liver cancer is a serious detrimental condition. Addressing the challenges associated with metastatic liver cancer requires a comprehensive understanding of its harmful nature and development of targeted treatment strategies.
Mechanisms underlying liver cancer metastasis are complex. In 2012, Biamonti et al[57] explored the role of AS in EMT, elucidating the link between AS and the invasive abilities of cancer cells. Understanding the effects of AS on EMT is critical for elucidating the underlying mechanisms of cancer metastasis and drug resistance. Next, a systematic review of liver transplantation in patients with liver metastases from neuroendocrine tumors highlighted the challenges posed by high recurrence rates, underscoring the need for precise patient selection and new treatment strategies[58]. Breakth
Metastatic liver cancer is a complex and multifaceted disease, and AS has been identified as a key factor in its progression. Several themes regarding the role of AS in metastatic liver cancer have emerged in the literature. AS is associated with EMT, a key process in cancer metastasis[57]. In addition, the splicing of specific genes such as CD44 has been shown to enhance the metastatic potential of cancer cells[61-63]. In addition, regulatory strategies to control AS in cancers, including metastatic liver cancer, remain largely unknown, suggesting gaps in our understanding of the underlying mechanisms[37,60]. In addition, associations between AS and metastatic phenotypes have been studied in various types of cancers, including colorectal and prostate cancers, suggesting that AS has a broader relevance in cancer metastasis[64,65]. Despite these insights, the existing studies of AS for metastatic liver cancer have some shortcomings. The functional mechanisms of AS in cancer, particularly liver metastasis, remain unclear[46,66]. Although a link between AS and cancer metastasis has been established, the specific regulatory procedures governing this process remain unclear[60]. In addition, the literature highlights the disappointing outcomes of liver transplantation for both primary and metastatic liver cancers, suggesting a lack of effective treatment strategies for metastatic liver disease[67]. This finding suggests that further research is needed to develop new treatment options for metastatic liver cancer. However, significant gaps exist in our understanding of the functional mechanisms and regulatory processes involved in AS in metastatic liver cancer. Addressing these gaps is critical for developing effective interventions for this challenging disease. This article is the first review of variable splicing in metastatic liver cancer, with the hope of providing new directions for future research.
In recent years, the importance of variable splicing in the development of liver metastases has been increasingly recognized. These breakthroughs underscore the potential of targeting AS events and related molecular pathways to inhibit the development and progression of metastatic cancers. Further research and clinical studies are essential to translate these findings into effective treatments for patients with metastatic liver cancer.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yoshinaga K, Japan S-Editor: Zhang H L-Editor: A P-Editor: Yuan YY
| 1. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Author Correction: Hepatocellular carcinoma. Nat Rev Dis Primers. 2024;10:10. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Li X, Ramadori P, Pfister D, Seehawer M, Zender L, Heikenwalder M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer. 2021;21:541-557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 190] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 3. | Naumann RW, Coleman RL, Brown J, Moore KN. Corrigendum to "Phase III trials in ovarian cancer: The evolving landscape of front line therapy" [Gynecol. Oncol. 153 (2019) 436-444]. Gynecol Oncol. 2020;156:512-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Li J, Zhu H, Sun L, Xu W, Wang X. Prognostic value of site-specific metastases in lung cancer: A population based study. J Cancer. 2019;10:3079-3086. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 5. | Kawai H, Shiba H, Kanehira M, Sakamoto T, Furukawa K, Yanaga K. Successful resection of a solitary metastatic liver tumor from prostate cancer 15 years after radical prostatectomy: a case report. Surg Case Rep. 2017;3:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 6. | Honkanen TJ, Luukkainen MEK, Tikkanen A, Karihtala P, Mäkinen M, Väyrynen JP, Koivunen JP. Immune cell profiles of metastatic HER2-positive breast cancer patients according to the sites of metastasis. Breast Cancer Res Treat. 2022;191:443-450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 7. | Yoshida T, Matsue H, Okazaki N, Yoshino M. Ultrasonographic differentiation of hepatocellular carcinoma from metastatic liver cancer. J Clin Ultrasound. 1987;15:431-437. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976-986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 325] [Cited by in F6Publishing: 370] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 9. | Melichar B, Voboril Z, Cerman J Jr, Melicharová K, Nozicka J, Mergancová J, Voboril R, Jandík P. Hepatic arterial infusion chemotherapy in gastric cancer: a report of four cases and analysis of the literature. Tumori. 2004;90:428-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Doi H, Beppu N, Kitajima K, Kuribayashi K. Stereotactic Body Radiation Therapy for Liver Tumors: Current Status and Perspectives. Anticancer Res. 2018;38:591-599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Urbanski LM, Leclair N, Anczuków O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA. 2018;9:e1476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 205] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 12. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 619] [Cited by in F6Publishing: 712] [Article Influence: 101.7] [Reference Citation Analysis (2)] |
| 13. | Bacalbaşa N, Alexandrescu ST, Popescu I. A role for hepatic surgery in patients with liver metastatic breast cancer: review of literature. Hepat Oncol. 2015;2:159-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 14. | Bradley RK, Anczuków O. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer. 2023;23:135-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 47] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 15. | Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 502] [Cited by in F6Publishing: 436] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 16. | Jimenez M, Arechederra M, Ávila MA, Berasain C. Splicing alterations contributing to cancer hallmarks in the liver: central role of dedifferentiation and genome instability. Transl Gastroenterol Hepatol. 2018;3:84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Soto M, Reviejo M, Al-Abdulla R, Romero MR, Macias RIR, Boix L, Bruix J, Serrano MA, Marin JJG. Relationship between changes in the exon-recognition machinery and SLC22A1 alternative splicing in hepatocellular carcinoma. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Reviejo M, Soto M, Lozano E, Asensio M, Martínez-Augustin O, Sánchez de Medina F, Marin JJG. Impact of alternative splicing on mechanisms of resistance to anticancer drugs. Biochem Pharmacol. 2021;193:114810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Marin JJG, Reviejo M, Soto M, Lozano E, Asensio M, Ortiz-Rivero S, Berasain C, Avila MA, Herraez E. Impact of Alternative Splicing Variants on Liver Cancer Biology. Cancers (Basel). 2021;14:18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 515] [Cited by in F6Publishing: 526] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 21. | Chen H, Gao F, He M, Ding XF, Wong AM, Sze SC, Yu AC, Sun T, Chan AW, Wang X, Wong N. Long-Read RNA Sequencing Identifies Alternative Splice Variants in Hepatocellular Carcinoma and Tumor-Specific Isoforms. Hepatology. 2019;70:1011-1025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 22. | Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 570] [Cited by in F6Publishing: 582] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 23. | Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 131] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 24. | Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635-2641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 266] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 25. | Lee SE, Alcedo KP, Kim HJ, Snider NT. Alternative Splicing in Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol. 2020;10:699-712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 26. | Caruso JA, Carruthers NJ, Thibodeau B, Geddes TJ, Dombkowski AA, Stemmer PM. Global Signaling Profiling in a Human Model of Tumorigenic Progression Indicates a Role for Alternative RNA Splicing in Cellular Reprogramming. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 27. | Bohnsack KE, Yi S, Venus S, Jankowsky E, Bohnsack MT. Cellular functions of eukaryotic RNA helicases and their links to human diseases. Nat Rev Mol Cell Biol. 2023;24:749-769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 20] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 28. | Kumar D, Das M, Oberg A, Sahoo D, Wu P, Sauceda C, Jih L, Ellies LG, Langiewicz MT, Sen S, Webster NJG. Hepatocyte Deletion of IGF2 Prevents DNA Damage and Tumor Formation in Hepatocellular Carcinoma. Adv Sci (Weinh). 2022;9:e2105120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Clapéron A, Chrétien Y, Rey C, Scatton O, Soubrane O, Conti F, Praz F, Housset C, Rosmorduc O, Desbois-Mouthon C. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974-3986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 30. | Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647-7654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 492] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 31. | Farina AR, Cappabianca L, Sebastiano M, Zelli V, Guadagni S, Mackay AR. Hypoxia-induced alternative splicing: the 11th Hallmark of Cancer. J Exp Clin Cancer Res. 2020;39:110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 32. | Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, Rauch J, Kolch W, Zender L, Karni R. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA. 2014;20:505-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 33. | Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064-1074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 419] [Cited by in F6Publishing: 467] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 34. | Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 416] [Cited by in F6Publishing: 430] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 35. | Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927-939. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2783] [Cited by in F6Publishing: 2885] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 36. | Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818-829. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2157] [Cited by in F6Publishing: 2267] [Article Influence: 141.7] [Reference Citation Analysis (0)] |
| 37. | Lee J, Pang K, Kim J, Hong E, Lee J, Cho HJ, Park J, Son M, Park S, Lee M, Ooshima A, Park KS, Yang HK, Yang KM, Kim SJ. ESRP1-regulated isoform switching of LRRFIP2 determines metastasis of gastric cancer. Nat Commun. 2022;13:6274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 8] [Reference Citation Analysis (0)] |
| 38. | Bhattacharya R, Mitra T, Ray Chaudhuri S, Roy SS. Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J Cell Biochem. 2018;119:3373-3383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 39. | Ueda J, Matsuda Y, Yamahatsu K, Uchida E, Naito Z, Korc M, Ishiwata T. Epithelial splicing regulatory protein 1 is a favorable prognostic factor in pancreatic cancer that attenuates pancreatic metastases. Oncogene. 2014;33:4485-4495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 40. | Walser TC, Jing Z, Tran LM, Lin YQ, Yakobian N, Wang G, Krysan K, Zhu LX, Sharma S, Lee MH, Belperio JA, Ooi AT, Gomperts BN, Shay JW, Larsen JE, Minna JD, Hong LS, Fishbein MC, Dubinett SM. Silencing the Snail-Dependent RNA Splice Regulator ESRP1 Drives Malignant Transformation of Human Pulmonary Epithelial Cells. Cancer Res. 2018;78:1986-1999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Chen QF, Li W, Wu P, Shen L, Huang ZL. Alternative splicing events are prognostic in hepatocellular carcinoma. Aging (Albany NY). 2019;11:4720-4735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 42. | Deng K, Yao J, Huang J, Ding Y, Zuo J. Abnormal alternative splicing promotes tumor resistance in targeted therapy and immunotherapy. Transl Oncol. 2021;14:101077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Lu ZX, Huang Q, Park JW, Shen S, Lin L, Tokheim CJ, Henry MD, Xing Y. Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol Cancer Res. 2015;13:305-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 44. | Jbara A, Lin KT, Stossel C, Siegfried Z, Shqerat H, Amar-Schwartz A, Elyada E, Mogilevsky M, Raitses-Gurevich M, Johnson JL, Yaron TM, Ovadia O, Jang GH, Danan-Gotthold M, Cantley LC, Levanon EY, Gallinger S, Krainer AR, Golan T, Karni R. RBFOX2 modulates a metastatic signature of alternative splicing in pancreatic cancer. Nature. 2023;617:147-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 13] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 45. | Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, Wellner U, Hopt UT, Brummer T, Brabletz S, Brabletz T, Stemmler MP. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer. 2015;137:2566-2577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 133] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 46. | Liu P, Wang Z, Ou X, Wu P, Zhang Y, Wu S, Xiao X, Li Y, Ye F, Tang H. The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer. 2022;21:198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 84] [Reference Citation Analysis (0)] |
| 47. | Chiou SH, Risca VI, Wang GX, Yang D, Grüner BM, Kathiria AS, Ma RK, Vaka D, Chu P, Kozak M, Castellini L, Graves EE, Kim GE, Mourrain P, Koong AC, Giaccia AJ, Winslow MM. BLIMP1 Induces Transient Metastatic Heterogeneity in Pancreatic Cancer. Cancer Discov. 2017;7:1184-1199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 48. | Krishnamachary B, Penet MF, Nimmagadda S, Mironchik Y, Raman V, Solaiyappan M, Semenza GL, Pomper MG, Bhujwalla ZM. Hypoxia regulates CD44 and its variant isoforms through HIF-1α in triple negative breast cancer. PLoS One. 2012;7:e44078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 49. | Han J, Li J, Ho JC, Chia GS, Kato H, Jha S, Yang H, Poellinger L, Lee KL. Hypoxia is a Key Driver of Alternative Splicing in Human Breast Cancer Cells. Sci Rep. 2017;7:4108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 50. | Memon D, Dawson K, Smowton CS, Xing W, Dive C, Miller CJ. Hypoxia-driven splicing into noncoding isoforms regulates the DNA damage response. NPJ Genom Med. 2016;1:16020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | Kemmerer K, Weigand JE. Hypoxia reduces MAX expression in endothelial cells by unproductive splicing. FEBS Lett. 2014;588:4784-4790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 52. | Bowler E, Porazinski S, Uzor S, Thibault P, Durand M, Lapointe E, Rouschop KMA, Hancock J, Wilson I, Ladomery M. Hypoxia leads to significant changes in alternative splicing and elevated expression of CLK splice factor kinases in PC3 prostate cancer cells. BMC Cancer. 2018;18:355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 53. | Rajapaksha I. Liver Fibrosis, Liver Cancer, and Advances in Therapeutic Approaches. Livers. 2022;2:372-386. [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 54. | Li W, Deng R, Liu S, Wang K, Sun J. Hepatitis B virus-related hepatocellular carcinoma in the era of antiviral therapy: The emerging role of non-viral risk factors. Liver Int. 2020;40:2316-2325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 55. | Wang X, Yu GY, Chen M, Wei R, Chen J, Wang Z. Pattern of distant metastases in primary extrahepatic bile-duct cancer: A SEER-based study. Cancer Med. 2018;7:5006-5014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 56. | Ji Y, Zhu J, Zhu L, Zhu Y, Zhao H. High-Intensity Focused Ultrasound Ablation for Unresectable Primary and Metastatic Liver Cancer: Real-World Research in a Chinese Tertiary Center With 275 Cases. Front Oncol. 2020;10:519164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 57. | Biamonti G, Bonomi S, Gallo S, Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell Mol Life Sci. 2012;69:2515-2526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Moris D, Tsilimigras DI, Ntanasis-Stathopoulos I, Beal EW, Felekouras E, Vernadakis S, Fung JJ, Pawlik TM. Liver transplantation in patients with liver metastases from neuroendocrine tumors: A systematic review. Surgery. 2017;162:525-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 59. | Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C; Cancer Genome Atlas Research Network, Rätsch G. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell. 2018;34:211-224.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 451] [Cited by in F6Publishing: 483] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 60. | Fish L, Khoroshkin M, Navickas A, Garcia K, Culbertson B, Hänisch B, Zhang S, Nguyen HCB, Soto LM, Dermit M, Mardakheh FK, Molina H, Alarcón C, Najafabadi HS, Goodarzi H. A prometastatic splicing program regulated by SNRPA1 interactions with structured RNA elements. Science. 2021;372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 61. | Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H, Osawa T, Kanki Y, Minami T, Aburatani H, Ohmura M, Kubo A, Suematsu M, Takahashi K, Saya H, Nagano O. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 280] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 62. | Louderbough JM, Schroeder JA. Understanding the dual nature of CD44 in breast cancer progression. Mol Cancer Res. 2011;9:1573-1586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 63. | Ouhtit A, Abd Elmageed ZY, Abdraboh ME, Lioe TF, Raj MH. In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am J Pathol. 2007;171:2033-2039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 64. | Munkley J, Li L, Krishnan SRG, Hysenaj G, Scott E, Dalgliesh C, Oo HZ, Maia TM, Cheung K, Ehrmann I, Livermore KE, Zielinska H, Thompson O, Knight B, McCullagh P, McGrath J, Crundwell M, Harries LW, Daugaard M, Cockell S, Barbosa-Morais NL, Oltean S, Elliott DJ. Androgen-regulated transcription of ESRP2 drives alternative splicing patterns in prostate cancer. Elife. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 65. | Torres S, García-Palmero I, Marín-Vicente C, Bartolomé RA, Calviño E, Fernández-Aceñero MJ, Casal JI. Proteomic Characterization of Transcription and Splicing Factors Associated with a Metastatic Phenotype in Colorectal Cancer. J Proteome Res. 2018;17:252-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 66. | Wang Z, Yang L, Wu P, Li X, Tang Y, Ou X, Zhang Y, Xiao X, Wang J, Tang H. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. 2022;21:29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 67. | Hackl C, Schlitt HJ, Kirchner GI, Knoppke B, Loss M. Liver transplantation for malignancy: current treatment strategies and future perspectives. World J Gastroenterol. 2014;20:5331-5344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 26] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |