Published online Dec 26, 2015. doi: 10.4330/wjc.v7.i12.843
Peer-review started: May 31, 2015
First decision: August 16, 2015
Revised: September 21, 2015
Accepted: October 12, 2015
Article in press: October 13, 2015
Published online: December 26, 2015
microRNAs (miRNAs) are powerful regulators of posttranscriptional gene expression and play an important role in pathophysiological processes. Circulating miRNAs can be quantified in body liquids and are promising biomarkers in numerous diseases. In cardiovascular disease miRNAs have been proven to be reliable diagnostic biomarkers for different disease entities. In cardiac fibrosis (CF) and heart failure (HF) dysregulated circulating miRNAs have been identified, indicating their promising applicability as diagnostic biomarkers. Some miRNAs were successfully tested in risk stratification of HF implementing their potential use as prognostic biomarkers. In this respect miRNAs might soon be implemented in diagnostic clinical routine. In the young field of miRNA based research advances have been made in identifying miRNAs as potential targets for the treatment of experimental CF and HF. Promising study results suggest their potential future application as therapeutic agents in treatment of cardiovascular disease. This article summarizes the current state of the various aspects of miRNA research in the field of CF and HF with reduced ejection fraction as well as preserved ejection fraction. The review provides an overview of the application of circulating miRNAs as biomarkers in CF and HF and current approaches to therapeutically utilize miRNAs in this field of cardiovascular disease.
Core tip: Recent study results suggest microRNAs (miRNAs) as promising biomarkers in the diagnosis of heart failure (HF) with reduced ejection fraction (HFrEF) and with preserve ejection fraction (HFpEF). The therapeutic application of antagomirs and mirmimics in heart failure is still in its infancy but promising experimental results are reported. This review provides an overview of miRNAs as diagnostic and prognostic biomarkers in HF and gives details on the utilization of miRNAs in the differentiated diagnosis of HFpEF and HFrEF. The manuscript evaluates the therapeutic applicability of miRNAs in HF and thus provides valuable information for researchers dealing with miRNAs in HF.
- Citation: Schulte C, Westermann D, Blankenberg S, Zeller T. Diagnostic and prognostic value of circulating microRNAs in heart failure with preserved and reduced ejection fraction. World J Cardiol 2015; 7(12): 843-860
- URL: https://www.wjgnet.com/1949-8462/full/v7/i12/843.htm
- DOI: https://dx.doi.org/10.4330/wjc.v7.i12.843
The term “microRNA” (miRNA) was established in 1993 when researchers started to study the function of small RNAs[1]. Lin-4 was the first miRNA described[2] and after its discovery scientists began to recognize miRNAs’ importance as regulators in gene expression. Ever since miRNAs have not only been assessed for their promising regulatory role in various diseases, but also their diagnostic potential in risk prediction as well as their use as circulating biomarkers[3-5]. Furthermore, promising data have depicted miRNAs as gene specific therapeutic targets in disease modeling[6].
Numerous studies have analyzed miRNAs with respect to their utilization as disease-specific biomarkers. In cardiovascular disease, miRNAs have successfully been proven to be quantitatively modified in particular disease entities such as myocardial fibrosis and heart failure (HF)[7-12].
In this article we will review the value of miRNAs in cardiac fibrosis (CF) and HF. We will discuss the current knowledge about their role in two different entities of the disease - HF with reduced ejection fraction (HFrEF) and HF with preserved ejection fraction (HFpEF). In particular, we will provide an overview about the use of miRNAs as diagnostic and prognostic biomarkers in CF and HF as well as potential therapeutic agents.
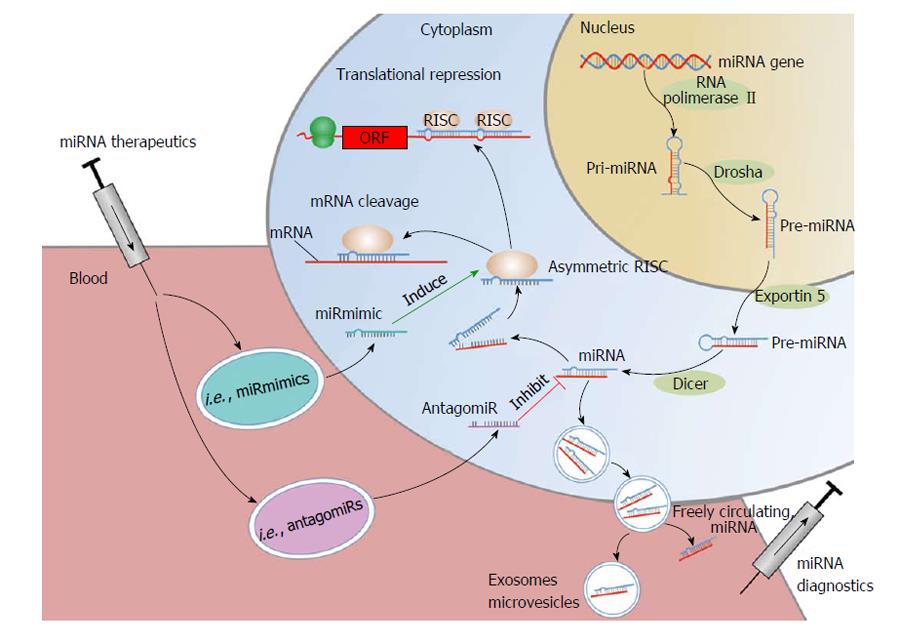
miRNAs are non-coding RNAs with a length of 19-25 nucleotides[13,14]. By binding to the 3’-untranslated region of target messenger RNAs (mRNAs), miRNA either initiate translational repression or degradation of mRNAs thereby regulating gene expression at the post-translational stage[15,16] (Figure 1). Every single miRNA has target sites in hundreds of different genes[17]. At the same time computational prediction of target mRNAs suggests that more than 60% of all mammalian protein-coding genes are conserved targets of miRNAs[18]. At the time of writing this review 2588 mature homo sapiens miRNAs were listed in “miRBase” (mirbase.org).
miRNA quantification showed organ- and cell-specific expression patterns of certain miRNAs[19] while quantification measures have shown concentration-dependent effects in pathologically altered organs[20]. In-vitro findings suggest groups of miRNAs being specifically up and down-regulated and polymorphisms in the miRNA regulatory pathway - so called miRSNPs - have been found to be associated with different types of disease[21-24]. miRNAs fulfill several criteria of an ideal biomarker: stability in the circulation, tissue- and pathology-specific regulation as well as high sensitivity and specificity. These characteristics predestine miRNAs as biomarkers. In fact, there is evidence that miRNAs’ applicability as circulating biomarkers for certain diseases might even exceed that of protein-based biomarkers[25,26]. The field of miRNA research has paved the way for the development of new means of biomarker-based risk stratification for cardiovascular events. In this regard promising data have been collected in large-scale prospective clinical studies[27].
The discovery of miRNAs as promising new biomarkers in cardiovascular disease has ignited great expectations and especially in the field of HF, the last years have witnessed great success. The syndrome of HF ranges among the leading causes of death and morbidity world-wide[28] with mortality rates of up to 50% in patients with new onset HF[29]. HF can be classified by the contraction of the left ventricle into HFrEF and HFpEF. While HFrEF is defined by a reduced left ventricular ejection fraction (LVEF), HFpEF describes HF patients with normal or only mildly reduced LVEF (over 50%)[30]. Approximately half of all HF patients present with preserved LVEF[31,32] illustrating its clinical importance, while morbidity and mortality are suggested to be equally distributed.
There are three major causes of HF: hypertensive heart disease, ischemic heart disease and idiopathic dilated cardiomyopathy. Hypertension initiates molecular pathways that lead to increased cardiomyocyte size and protein synthesis as well as augmented sarcomer organization[33,34]. Persisting hypertrophy is associated with an unfavorable outcome and can result in HF and sudden death[35,36]. Independently from the underlying pathology failing hearts remodel in regard to extracellular matrix and myocyte size. This leads to augmented hypertrophy and death of cardiomyocytes followed by tissue fibrosis and scarring[37]. CF results in increased myocardial stiffness affecting systolic as well as diastolic left ventricular function[37,38]. The initial molecular steps in the development of HF can hardly be analyzed using imaging techniques and protein biomarkers may be involved at later stages only[39]. In this respect the diagnosis of HF is most often been made at an advanced stage of the disease when symptoms and physical confinement have already developed. The fact that so-called early HFpEF usually is clinically apparent merely under exercise conditions complicates an early diagnosis[40-42]. This determines the clinical need for markers identifying the disease at the earliest possible stage.
Given that cardiac remodeling and fibrosis are significant factors in the development of ventricular wall stiffness with compromised ventricular contractility and compliance, the expression of miRNAs is directly linked to the development of HF - with preserved and with reduced ejection fraction. miRNAs were identified to play a major role in the transcriptional and translational changes in gene expression with respect to cardiac hypertrophy and fibrosis[36,43]. The regulatory involvement of miRNAs in the development of cardiac hypertrophy and fibrosis ultimately suggests their causal roll in HF.
In a mouse model of aortic constriction induced cardiac hypertrophy van Rooij et al[36] described altered levels of several miRNAs in murine cardiomyocytes while Sayed et al[44] found a set of more than 50 miRNAs dysregulated in a similar setting with induced hypertrophy. Especially miR-1 was identified as significantly down-regulated compared to sham operated controls, probably mediated via an inhibition of the translation of calmodulin-encoding mRNAs[45]. Similar results were reported by Carè et al[46]. They reported down-regulated levels of miR-1 and miR-133 in cardiomyocytes of hypertrophic murine as well as human hearts[46]. Furthermore, in an in-vitro model the authors found a causal relationship between adeno-virus induced elevation of miR-1 and miR-133 levels and an inhibition of cardiac hypertrophy. Supporting data were reported from an in-vitro study involving neonatal rat cardiomyocytes[47]. Recently miR-150 has been described as a regulator in cardiac hypertrophy[48]. In a mouse model the authors induced cardiac hypertrophy by aortic banding and found miR-150 levels down-regulated compared to sham operated animals[48]. Several more in-vitro and mouse model studies reported altered miRNA levels in cardiac hypertrophy on the one hand and induction of cardiac hypertrophy by artificial alterations of specific miRNAs on the other[49-52]. Especially miR-1[44,53], miR-21[54,55], miR-133[53,54], miR-195[36,56], miR-208[57-59] were proven to be involved in the regulation of cardiac hypertrophy. Alterations of miRNA levels were also reported in induced hypertrophy of cardiomyocytes in engineered heart tissue[60].
The heart’s initial hypertrophic response to volume overload or increased afterload as well as pathological conditions after myocardial infarction (MI) is followed by the process of remodeling which leads to CF. A major regulatory roll of miRNAs in the process of cardiac remodeling and fibrosis was suggested when a key enzymatic step towards miRNA activation catalyzed by the enzyme Dicer was blocked in knock out mice[61]. The authors found biventricular enlargement, myocyte hypertrophy and pronounced CF[61]. miRNAs involved in regulatory pathways of hypertrophy such as miR-208 have also been found in cardiac remodeling[57]. van Rooij et al[57] found mutant mice overexpressing miR-208 to not develop cardiomyocyte fibrosis despite being exposed to an increased afterload. Opposite, miR-208a has been identified as a regulator of endoglin expression and increases myocardial fibrosis in volume overloaded hearts[62]. Comparable results were reported in a model of cultured rat myoblasts[59].
miR-133a was reported to be down-regulated in a mouse model of aortic constriction-induced hypertrophy[63]. Expression of miR-133a prevented this down-regulation while the authors found less myocardial fibrosis along with improved diastolic function of the analyzed mouse hearts[63]. Similar results were reported from analyses of CF in canines. Shan et al[64] described reduced miR-133 and miR-590 levels in canine hearts after nicotine-induced CF mediated by up-regulation of transforming growth factor β (TGF-β) 1 and TGF-β RII. By transfection of miR-133 or miR-590 into cultured atrial fibroblasts it was possible to reduce fibroblast activity as well as collagen production while this effect was reversible by administration of antisense oligonucleotides against miR-133 or miR-590[64]. Confirming results of reducing CF by miR-133 induction were recently reported in a mouse model of aortic banding-induced hypertrophy[65]. Besides miR-133 also miR-30 was reported to control pro-fibrotic proteins and thus regulate changes in the extracellular matrix of the myocardium[66]. The decrease of miR-133 and miR-30 in a gene-modulated rat model of pathological cardiac hypertrophy was found to be linked with an up-regulation of collagen synthesis and CF[66].
In a transgenic mouse model miR-21 was discovered as a key regulator of signaling pathways in cardiac fibroblasts controlling the extent of cardiac hypertrophy and interstitial fibrosis[55]. A different working group was able to show that elevated miR-21 expression was highly related to CF[67]. These findings are in line with the observation that miR-21 is involved in the regulation pathway of cardiac fibroblasts in infarcted mouse hearts[68]. Cardin et al[69] were able to suppress atrial fibrosis in miR-21 knock out mice after induced MI. Also, the authors succeeded in depressing post infarction fibrosis by means of anti-miR-21 reduction of miR-21 availability. More recent studies validate these findings of miR-21 promoting cardiac remodeling and fibrosis[67,70,71]. On the other hand silencing of miR-21 by means of antagomirs resulted in cardiomyocyte necrosis and apoptosis[72,73] indicating the integration of this miRNA in cardiac remodeling.
MI is frequently followed by ventricular remodeling processes and fibrotic structural changes in the infarcted areas. This process eventually leads to HF. A miRNA involved in post-MI CF is miR-24. In a mouse model of induced MI Wang et al[74] reported miR-24 down-regulated and found a simultaneous increase of extracellular matrix remodeling. In-vivo lentivirus-based intramyocardial elevation of miR-24 levels caused attenuation of fibrosis in the infarct border zone[74]. The authors described TGF-β to mediate the miR-24 modulated effect and concluded miR-24 to be a potential target for the treatment of post-MI remodeling.
miR-29 is involved in fibrotic processes in different types of body tissue[75,76]. It controls a variety of pro-fibrotic genes such as collagens, fibrillins, laminins, integrins and elastin[77]. Furthermore, miR-29 negatively regulates a number of anti-apoptotic genes, including Tcl-1, Mcl-1, YY1, p85a, CDC42 and DNMT3[77-81]. In a mouse model of induced MI miR-29 was down-regulated[82]. Further analyses revealed miR-29 being predominantly expressed in cardiac fibroblasts and the in-vivo inhibition of miR-29 resulted in an induction of collagen mRNA expression[82]. Raising miR-29 concentrations using Mirmimics lead to a down-regulation of collagen mRNA expression[82]. Affirmative results were recently reported by Yang et al[83]. With respect to these results miR-29 can be attributed a key roll in regulation of tissue fibrosis and CF in particular.
Study results like these gave rise to projects analysing the roll of miRNAs in manifest HF.
In the process of HF development different intracellular signaling pathways are activated including an up-regulation of structural fetal genes, such as β-myosin heavy chain (β-MHC) and down-regulation of adult structural genes, such as α-MHC[84-86]. miRNAs are involved in regulatory processes of activating fetal genes that are known to be up-regulated in failing hearts[87]. The involvement of miR-208 in α- and β-MHC regulation was reported by van Rooij et al[57]. Via this mechanism miR-208 regulates cardiomyocytes growth under stress conditions as reported by the authors in a transgenic mouse model[57]. The same working group had previously found a set of miRNAs up-regulated not only in a mouse model of induced hypertrophy but also in failing human hearts[36]. They found an increased expression of miR-24, miR-125b, miR-195, miR-199a and miR-214 in both mice and human hearts and postulated that these miRNAs are part of a molecular signature of adverse cardiac remodeling[36].
Besides the development of cardiac hypertrophy and fibrosis as described above a knockout of the key enzyme Dicer, essential for intracellular miRNA processing, leads to dilatative cardiomyopathy (DCM) and HF[88]. The authors found reduced Dicer expression in human failing hearts and reported a significant increase of Dicer expression in hearts of patients with improved cardiac function after implantation of left ventricular assist device (LVAD) for HF[88]. These results depict the importance of miRNAs in the regulation of pathophysiologic processes involved in the development of HF and have lead to considerations of clinical implications of miRNAs dysregulated in cardiomyopathies and HF in particular. Matkovich et al[89] drove the Dicer-related findings by Chen et al[88] (mentioned above) a step further and analyzed a miRNA expression profile of cardiac tissue from HF patients with and without LVAD-based recovery compared to healthy controls. The authors found 28 miRNAs up-regulated in failing hearts compared to healthy controls and 20 of these miRNAs returned to near normal levels in the LVAD-treated group with significant improvement of left ventricular performance[89]. While these results link defined miRNAs to clinically apparent HF and suggest their potential in treatment monitoring, a more distinct analysis of miRNAs in different types of cardiomyopathies was performed by Ikeda et al[90]. In left ventricular biopsy samples of 67 humans with ischemic cardiomyopathy (ICM), DCM, aortic stenosis and healthy controls they analyzed miRNA expression[90]. Using a genome-wide miRNA expression profiling they detected 87 miRNAs and found their expression profiles significantly altered in the three heart diseases compared to healthy controls. While seven miRNAs were altered in the same direction in all three disease entities, the global pattern of miRNA expression was distinct in different types of HF[90]. miR-19 appeared as the most strongly down-regulated miRNA in DCM and AS but not in ICM, while miR-1 was down-regulated in all three diseases. miR-214 - considered pro-hypertrophic[36] - was most strongly up-regulated. Surprisingly, miR-133 and miR-208 levels, which are associated with myocardial hypertrophy[46,57-59,63] and fibrosis[59,62,64], were unchanged. These reported data suggest miRNAs to be specifically dysregulated in different types of HF pathology. In this regard an interesting study was recently reported by Leptidis et al[91] who performed miRNA deep sequencing analyses in myocardial biopsies of end stage DCM, hypertrophic cardiomyopathy (HCM) and healthy controls to analyze the human heart’s miRNOME with respect to these two different HF pathologies. They were able to identify a set of ten miRNAs (miR-23b, miR-30d, miR-125a, miR-143, miR-145, miR-193, miR-197, miR-342, miR-365, miR-455) that is differentially expressed in HCM and DCM compared to healthy controls and had not been linked to HF previously[91]. The authors were able to confirm previously described dysregulated levels of miR-133a, miR-1, miR-21, miR-214, miR-212, miR-29, miR-129, miR199a in HCM, while miR-119 and miR-214 expression was reported only to be altered in DCM[91]. miR-145 was identified as a new regulator of pathologic left-ventricular remodeling. Satoh et al[92] who analyzed miRNA expression in cardiac tissue from myocardial biopsies of patients with DCM reported higher levels of miR-208, miR-208b and miR-499 than in healthy controls. Follow-up revealed baseline miR-208 levels to be strong predictors of clinical outcome[92] indicating a potential utilization of miRNAs in risk prediction of HF.
Based on these results, the potential applicability of miRNAs as distinct biomarkers for the diagnosis of HF and for different entities of the disease seems possible. miRNAs seem to be promising biomarkers in risk prediction of HF patients. For obvious reasons, though, the availability of heart tissue is limited and therefore different sources of biomaterial for miRNA analysis are needed. In this respect body fluids appear to present an ideal origin to non-invasively win such biomaterial.
Besides regulating gene expression and phenotypic control in the cell of origin[93] and mediating metabolism on an intracellular level[94] miRNAs are also secreted from the producing cell and capable of transmitting their silencing signals to different cells[95]. miRNAs have been detected in numerous body fluids such as serum and plasma as well as saliva and urine[25,26] and can be found in pericardial fluid of HF patients[96]. Consequently, miRNAs have been tested to function as detectable extracellular messengers in cell-to-cell communication[97]. Their structure prevents miRNAs from early degradation in circulating blood[25,98-102] and their ideal biomarker characteristics including size, abundance and tissue specificity suggest circulating miRNAs as blood-based biomarkers for tissue injury[12,103-105].
The ability to detect and measure miRNAs in a minimal-invasive way has led to their evaluation as potential circulating biomarkers for cardiovascular disease[3,106]. The promising results of disease-specific cellular miRNA dysregulation in HF and their suitable characteristics with regard to circulating biomarker diagnostics have led to their evaluation as blood based biomarkers in HF. In a rat model of induced left ventricular hypertrophy with consecutive development of HF the authors reported significantly elevated plasma levels of miR-16, miR-20b, miR-93, miR-106b, miR-223 and miR-423-5p[107]. These results were in line with earlier findings of Tijsen et al[108] who were amongst the first researchers to evaluate circulating miRNAs as diagnostic biomarkers in HF in a clinical approach. The authors reported that besides 6 miRNAs (miR-18ba, miR-129-5p, miR-1254, miR-675, oncomir HS_202.1 miR-622), that were moderately elevated in plasma of 30 HF patients compared to 20 dyspnea patients and 20 healthy controls, miR-423-5p was found to be a significant predictor of HF diagnosis in a multivariate logistic regression model[108]. Despite the small sample size and suboptimal matching of baseline characteristics[109] these were promising initial results that were confirmed by Goren et al[110] in a similar study setup. The authors were one of the first groups to perform a screening of circulating miRNAs on a larger scale in cardiovascular disease and HF. They screened 186 miRNAs in serum of 30 HF patients compared to 30 healthy controls and were able to detect four miRNAs (miR-423-5p, miR-320a, miR-22, miR-92b) that were up-regulated in the serum of HF patients compared to the control group[110]. Furthermore, the authors succeeded in generating a score out of these miRNAs that discriminates HF patients from healthy controls. The group was able to describe a significant association between the miRNA score and several established prognostic HF parameters such as NT-proBNP, a wide QRS complex and left ventricular (LV) dilatation underlining the significance of these results not only with respect to diagnostic but also to prognostic applicability of circulating miRNAs[110]. At that time the analyzed combination of miRNAs miR-320 and miR-423-5p had previously been associated with HF[86,111,112].
Ellis et al[113] analyzed miRNA plasma levels of 44 HF patients compared to 32 Chronic obstructive pulmonary disease (COPD) patients, 59 patients with breathlessness for other diagnoses and 15 healthy controls after an initial miRNA screening phase. Not only were seven miRNAs (miR-103, miR-142-3p, miR-342-3p, miR-199a, miR-23a, miR-27b, miR-324-5p) associated with the diagnosis of HF in regression and receiver operating characteristics (ROC) analysis, plasma levels of four miRNAs (miR-103, miR-142-3p, miR-30b and miR-342-3p) were able to distinguish between HF and exacerbation of COPD, other causes of dyspnea and controls[113]. Although miR-423-5p could not be identified as a predictor of HF diagnosis, the addition of miR-423-5p to NT-proBNP significantly improved the area under the operating receiver curve (AUC) for predicting the diagnosis HF[113]. These findings confirm previous results of the potential applicability of miR-423-5p as circulating biomarker in HF diagnosis.
In a larger clinical trial serum miRNA levels of 81 HF patients were compared to 60 non-HF patients and 15 healthy subjects[114]. The authors reported a set of 24 miRNAs significantly down-regulated in the HF group compared to controls. miR-26b-5p, miR-145-5p, miR-92a-3p, miR-30e-5p and miR-29a-3p inversely correlated with NT-proBNP and directly correlated with EF, while ROC analysis to predict differentiation of HF patients from non-HF cases revealed strong AUC values between 0.84 and 0.91, suggesting these miRNAs to be potentially strong circulating biomarkers in the diagnosis of HF[114].
Recently, Wong et al[115] identified miR-1233, miR-183-3p, miR-190a, miR-193b-3p, miR-193b-5p, miR-211-5p, miR-494 and miR-671-5p to be able to distinguish HF from healthy controls in plasma levels of 60 HF patients and 30 healthy subjects.
Circulating blood cells and endothelial cells contain higher miRNA concentrations than serum and plasma[116]. Thus, further approaches in quantification efforts of circulating miRNAs with respect to HF were aimed at analyzing their concentration in circulating blood cells such as peripheral blood mononuclear cells (PBMCs). Gupta et al[117] analyzed miRNA concentrations in PBMCs of 44 DCM HF patients compared to 48 healthy controls. Real time polymerase chain reaction (RT-PCR) revealed miR-548c and miR-548i significantly down-regulated in PBMCs of DCM patients, while miR-138 was up-regulated in PBMCs of those patients. ROC analysis showed an AUC of 0.85 for miR-548c with respect to its discriminatory power to distinguish DCM from controls[117].
Frequently, HF consecutively develops after ischemic events such as MI. Corsten et al[10] analyzed whole blood samples, plasma and urine of 32 acute myocardial infarction (AMI) patients compared to 36 non-AMI controls and reported plasma miR-208b and miR-499 to correlate with cardiac injury markers and, hence, to correlate with myocardial damage. miR-499 was significantly up-regulated in a subgroup of patients with acute HF. Another group that analyzed a predefined set of circulating miRNAs in plasma of 12 post-MI patients compared to 12 healthy controls was able to find levels of miR-1, miR-21, miR-29a, miR-133a and miR-208 altered in the time course after MI[118]. These miRNAs had previously been described to affect myocardial growth, hypertrophy, fibrosis and viability[118] implying that the same miRNAs that have been shown to be associated with these pathophysiological processes preceding HF can be found dysregulated in plasma of patients with cardiovascular disease.
Independently from the pathophysiological cause miRNAs have been reported to complement biomarker-based prediction of outcome in HF. In a study including 20 clinically stable and 22 decompensated HF patients as well as 15 healthy controls the authors performed a microarray-based miRNA profiling and reported a large number of miRNAs to be quantitatively dysregulated in HF patients compared to controls[119]. More importantly, Cox regression identified miR-182 to be able to predict cardiovascular mortality. Remarkably, the prognostic value of miR-182 was identified to be superior to NT-proBNP as well as high-sensitive C-reactive protein by ROC analysis[119]. Table 1 gives an overview of cellular and circulating miRNAs dysregulated in heart failure.
| miRNA | Study type | Bio-material | Group/size | Detection method | Effect | Value as biomarker | Ref. |
| Single miRNAs | |||||||
| Let-7 | Clinical | Tissue | ICM n = 10 | qRT-PCR | Up-regulated in DCM and ICM | Diagnostic | [90] |
| DCM n = 25 | |||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-1 | Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in DCM | Diagnostic | [90] |
| DCM n = 25 | |||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-15b | Clinical | Tissue | ICM n = 10 | qRT-PCR | Up-regulated in DCM | Diagnostic | [90] |
| DCM n = 25 | |||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-16 | Experimental | Plasma | Rats, hypertension-induced HF | qRT-PCR | Up-regulated in HF | Diagnostic | [107] |
| miR-17-5p | Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in DCM | Diagnostic | [90] |
| DCM n = 25 | |||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-18b | Clinical | Plasma | HF n = 30 | qRT-PCR | Up-regulated in HF | Diagnostic | [108] |
| Dyspnea n = 20 | |||||||
| HC n = 20 | |||||||
| miR-20b | Experimental | Plasma | Rats, hypertension-induced HF | qRT-PCR | Up-regulated in HF | Diagnostic | [107] |
| miR-211 | Clinical | Tissue | LVAD-patients | Micro-array | Up-regulated in HF | Experimental | [89] |
| miR-22 | Clinical | Serum | HFrEF n = 30 | qRT-PCR | Up-regulated in HFrEF | Diagnostic/ | [110] |
| HC n = 30 | prognostic | ||||||
| miR-23a | Clinical | Tissue | LVAD-patients | Micro-array | Up-regulated in HF | Experimental | [89] |
| miR-24 | Experimental | Tissue | Mice, human hearts | Micro-array | Up-regulated in HF, CH | Experimental | [36] |
| miR-26b-5p | Clinical | Plasma | HF n = 81 | qRT-PCR | Down-regulated | Diagnostic | [114] |
| HC n = 15 | |||||||
| miR-28 | Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in DCM | Diagnostic | [90] |
| DCM n = 25 | |||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-29a-3p | Clinical | Plasma | HF n = 81 | qRT-PCR | Down-regulated | Diagnostic | [114] |
| HC n = 15 | |||||||
| miR-30b | Clinical | Plasma | HF n = 44 | RT-PCR | Down-regulated in HF | Diagnostic | [113] |
| COPD n = 32 | |||||||
| Dyspnea n = 59 | |||||||
| HC n = 15 | |||||||
| miR-30e-5p | Clinical | Plasma | HF n = 81 | qRT-PCR | Down-regulated | Diagnostic | [114] |
| HC n = 15 | |||||||
| miR-92b | Clinical | Serum | HFrEF n = 30 | qRT-PCR | Up-regulated in HFrEF | Diagnostic/ | [110] |
| HC n = 30 | prognostic | ||||||
| miR-92a-3p | Clinical | Plasma | HF n = 81 | qRT-PCR | Down-regulated | Diagnostic | [114] |
| HC n = 15 | |||||||
| miR-93 | Experimental | Plasma | Rats, hypertension-induced HF | qRT-PCR | Up-regulated in HF | Diagnostic | [107] |
| miR-103 | Clinical | Plasma | HF n = 44 | RT-PCR | Down-regulated in HF | Diagnostic | [113] |
| COPD n = 32 | |||||||
| Dyspnea n = 59 | |||||||
| HC n = 15 | |||||||
| miR-106a | Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in DCM | Diagnostic | [90] |
| DCM n = 25 | |||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-106b | Experimental | Plasma | Rats, hypertension-induced HF | qRT-PCR | Up-regulated in HF | Diagnostic | [107] |
| miR-125b | Experimental | Tissue | Mice, human hearts | Micro-array | Up-regulated in HF, CH | Experimental | [36] |
| miR-126 | Clinical | Plasma | HF n = 10 | qRT-PCR | Down-regulated in HF | Diagnostic | [150] |
| HC n = 17 | |||||||
| miR-133 | Clinical | Tissue | LVAD-patients | Micro-array | Up-regulated in HF | Experimental | [89] |
| miR-138 | Clinical | PBMC | DCM n = 44 | qRT-PCR | Up-regulated in DCM | Diagnostic | [117] |
| HC n = 48 | |||||||
| miR-142-3p | Clinical | Plasma | HF n = 44 | RT-PCR | Down-regulated in HF | Diagnostic | [113] |
| COPD n = 32 | |||||||
| Dyspnea n = 59 | |||||||
| HC n = 15 | |||||||
| Clinical | Plasma | HFpEF n = 8 | qRT-PCR | Down-regulated in stable and | Diagnostic | [121] | |
| Stable DCM n = 10 | decompensated DCM | ||||||
| Decompensated DCM n = 13 | |||||||
| HC n = 8 | |||||||
| miR-145-5p | Clinical | Plasma | HF n = 81 | qRT-PCR | Down-regulated | Diagnostic | [114] |
| HC n = 15 | |||||||
| miR-182 | Clinical | Serum | HF n = 42 | Micro-array | Up-regulated in HF | Prognostic | [119] |
| HC n = 15 | |||||||
| miR-183-3p1 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Down-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-190a | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Down-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-193b-3p1 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Down-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-193b-5p1 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Down-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-195 | Experimental | Tissue | Mice, human hearts | Micro-array | Up-reglated in HF, CH | Experimental | [36] |
| Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in DCM | Diagnostic | [90] | |
| DCM n = 25 | and ICM | ||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-199a | Experimental | Tissue | Mice, human hearts | Micro-array | Up-reglated in HF, CH | Experimental | [36] |
| Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in | Diagnostic | [90] | |
| DCM n = 25 | DCM and ICM | ||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-208 | Experimental | Tissue | Mice | Micro-array | Up-regulated in HF, CF, CH | Experimental | [57] |
| Clinical | Tissue | DCM n = 82 | qRT-PCR | Up-regulated in DCM | Diagnostic/ | [92] | |
| HC n = 21 | prognostic | ||||||
| miR-208b | Clinical | Tissue | DCM n = 82 | qRT-PCR | Up-regulated in DCM | Diagnostic | [92] |
| HC n = 21 | |||||||
| miR-211-5p1 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Down-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-214 | Experimental | Tissue | Mice, human hearts | Micro-array | Up-reglated in HF, CH | Experimental | [36] |
| miR-222 | Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in | Diagnostic | [90] |
| DCM n = 25 | DCM and ICM | ||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-223 | Experimental | Plasma | Rats, hypertension-induced HF | qRT-PCR | Up-regulated in HF | Diagnostic | [107] |
| miR-320a | Clinical | Serum | HFrEF n = 30 | qRT-PCR | Up-regulated in HFrEF | Diagnostic/ | [110] |
| HC n = 30 | prognostic | ||||||
| miR-342-3p | Clinical | Plasma | HF n = 44 | RT-PCR | Down-regulated in HF | Diagnostic | [113] |
| COPD n = 32 | |||||||
| Dyspnea n = 59 | |||||||
| HC n = 15 | |||||||
| miR-422b | Clinical | Tissue | ICM n = 10 | qRT-PCR | Down-regulated in | Diagnostic | [90] |
| DCM n = 25 | DCM and ICM | ||||||
| AS n = 13 | |||||||
| HC n = 10 | |||||||
| miR-423-5p | Experimental | Plasma | Rats, hypertension-induced HF | qRT-PCR | Up-regulated in HF | Diagnostic | [107] |
| Clinical | Plasma | HF n = 30 | qRT-PCR | Up-regulated in HF | Diagnostic/ | [108] | |
| Dyspnea n = 20 | prognostic | ||||||
| HC n = 20 | |||||||
| Clinical | Serum | HFrEF n = 30 | qRT-PCR | Up-regulated in HFrEF | Diagnostic/ | [110] | |
| HC n = 30 | prognostic | ||||||
| Clinical | Plasma | HF n = 44 | RT-PCR | Down-regulated in HF | Prognostic when | [113] | |
| COPD n = 32 | combined with | ||||||
| Dyspnea n = 59 | NT-proBNP | ||||||
| HC n = 15 | |||||||
| miR-4941 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Down-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-499 | Clinical | Tissue | DCM n = 82 | qRT-PCR | Up-regulated in DCM | Diagnostic | [92] |
| HC n = 21 | |||||||
| Clinical | Plasma | Acute HF n = 33 | qRT-PCR | Up-regulation in acute HF | Diagnostic | [10] | |
| HC n = 34 | |||||||
| miR-548c | Clinical | PBMC | DCM n = 44 | qRT-PCR | Down-regulated in DCM | Diagnostic | [117] |
| HC n = 48 | |||||||
| miR-548i | Clinical | PBMC | DCM n = 44 | qRT-PCR | Down-regulated in DCM | Diagnostic | [117] |
| HC n = 48 | |||||||
| miR-671-5p1 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Up-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miR-675 | Clinical | Plasma | HF n = 30 | qRT-PCR | Up-regulated in HF | Diagnostic | [108] |
| Dyspnea n = 20 | |||||||
| HC n = 20 | |||||||
| miR-12331 | Clinical | Plasma | HF n = 60 (HFpEF n = 30; HFrEF n = 30) | qRT-PCR | Up-regulated in HF | Diagnostic | [115] |
| HC n = 28 | |||||||
| miRNA signatures | |||||||
| miR-520d-5p | Clinical | Whole blood | HFrEF n = 53 | qRT-PCR | Dysregulated in HF - superior to | Diagnostic | [120] |
| miR-558 | HC n = 39 | single miRNAs | |||||
| miR-122* | |||||||
| miR-200b* | |||||||
| miR-622 | |||||||
| miR-519e* | |||||||
| miR-1231 | |||||||
| miR-1228* | |||||||
The above studies suggest blood-based circulating miRNAs as potential strong tools in the diagnosis and risk evaluation of HF. On the other hand, most trials included rather small sample sizes and most identified miRNAs were not confirmed in repetitive studies.
Combining two or more biomarkers as a defined set for diagnostic purposes can enhance discriminatory power compared to the use of single biomarkers. In the field of miRNA biomarker research the assessment of sets (so-called signatures) of miRNAs might deliver superior results compared with the application of single miRNAs. In order to assess circulating miRNAs as biomarkers for HFrEF, Vogel et al[120] performed miRNA quantification measures in whole blood samples of 53 HFrEF patients with non-ischemic HF compared to 39 healthy controls. In a two-step screening-validation study the authors found a signature of eight miRNAs (miR-520d-5p, miR-558, miR-122*, miR-200b*, miR-622, miR-519e*, miR-1231 and miR-1228*) which reliably predicted the diagnosis of HFrEF with an AUC of 0.81[120] (Table 1). Compared to the most powerful single miRNAs miR-558, miR-122a, and miR-520d-5p (AUC between 0.7 and 0.71) this miRNA signature further improved discrimination of HFrEF patients from controls[120] confirming the idea to improve sensitivity and specificity when utilizing combinations of more than one miRNA.
Pathophysiologically, HFpEF is the clinical manifestation of LV diastolic dysfunction as a major differentiating factor from HFrEF. Therefore, diastolic dysfunction appears as a useful parameter in the early diagnosis of HFpEF.
Initial results proving the involvement of miR-21 in cardiac remodeling and fibrosis[55,67-73] (see above) suggested its roll in HFrEF. These findings were taken up by Dong et al[67] in order to analyze this miRNA with respect to HFpEF. The authors created a rat model of aortic constriction-induced HFpEF. HFpEF was diagnosed via echocardiographic parameters and quantitative RT-PCR (qRT-PCR) analyses showed higher cellular miR-21 levels in HFpEF rats compared to healthy controls[67]. These results confirm the former pathophysiologic miR-21 findings and indicate their potential to be transferrable to a functional level in HFpEF. In order to assess whether circulating miRNAs as opposed to cellular miRNAs can be utilized as biomarkers in the detection of HFpEF and in a differentiated diagnosis compared to HFrEF, results were published recently by Nair et al[121]. They analyzed miRNA plasma levels of HF patients with diastolic dysfunction. The authors found miR-454, miR-500 (both down-regulated) and miR-1246 (up-regulated) significantly dysregulated in diastolic dysfunction indicating that circulating miRNAs can serve as biomarkers for diastolic dysfunction[121]. This suggests itself to further considerations for miRNA-based diagnostics to differentiate HFrEF from HFpEF. Wong et al[115] performed a miRNA quantification of whole blood and plasma samples in 39 HFrEF and 19 HFpEF patients as well as 28 healthy controls and identified 344 miRNAs dysregulated between the three groups. Of these, 90 serum derived miRNAs were identified that showed high correlation with or an AUC > 0.7 for LVEF. Again a selection of 32 miRNAs with considerably high detection levels was made. These analytical steps allowed for a qualitative selection of promising miRNAs and those, that can easily be detected in serum. Further analyses of these 32 miRNAs in plasma of an independent cohort of 30 HFrEF and 30 HFpEF patients as well as 30 controls identified 12 miRNAs that could segregate HFrEF and HFpEF from non-HF controls as well as HFrEF from HFpEF[115]. In detail, miR-125a-5p, miR-183-3p, miR-193b-3p, miR-211-5p, miR-494, miR-638 and miR-671-5p differed significantly between HFrEF and controls while miR-1233, miR-183-3p, miR-190a, miR-193b-3p, miR-193b-5p and miR-545-5p showed significant differences in expression between HFpEF and controls[115]. miR-125a-5p (up-regulated in HFrEF - normal in HFpEF), miR-190a (down-regulated in HFpEF - normal in HFrEF), miR-550a-5p (directionally opposite expression pattern between HFrEF and HFpEF) and miR-638 (down-regulated in HFrEF - normal in HFpEF) were revealed to distinguish between HFrEF and HFpEF. Clinically relevant, the combinatory use of NT-proBNP with miR-125a-5p improved the AUC value to differentiate HFrEF from HFpEF from 0.83 for NT-proBNP alone to 0.91 for the combinatory use and thus significantly increased NT-proBNP’s discriminative diagnostic abilities[115]. Another aspect that was addressed in this study was the application of panels of miRNAs. The authors reported that miRNA panels had comparable performance to NT-proBNP with respect to the discrimination of HFrEF from HFpEF while single miRNAs tended to perform slightly inferior to NT-proBNP[115].
In order to identify a miRNA signature helping to differentiate HFpEF from HFrEF Watson et al[122] performed miRNA quantification analyses in sera of 90 HFpEF patients compared to 90 HFrEF patients and 90 healthy controls. The diagnosis of HFrEF and HFpEF was made echocardiographically. In an initial miRNA screening in serum samples of 15 individuals per group five candidate miRNAs (miR-30c, miR-146a, miR-221, miR-328 and miR-375) were identified as differentially expressed between the three groups and validated in an independent study cohort of 225 individuals[122]. The authors performed AUC analyses to differentiate HFpEF from HFrEF and reported an equal predictive value of any of the single miRNAs compared with the use of brain natriuretic peptide (BNP)[122]. Importantly, combinations of two or more of miR-146a, miR-221, miR-328 and miR-375 with BNP significantly improved the predictive power to differentiate HFpEF from HFrEF as compared with BNP alone in the AUC model[122]. The latter two very recent studies were the first to investigate circulating miRNAs as promising new biomarkers to differentiate HFpEF from HFrEF. They similarly provide evidence that combinatory utilization of miRNAs can improve discriminative power compared to single miRNAs. Critically evaluated, though, there was no overlap in the identified miRNAs in these studies that were able to distinguish between HFrEF and HFpEF. Schmitter et al[123] discussed potential explanations for these differences. The authors regarded several explanations relevant in this respect. First, methodological variances such as the choice of body liquid, detection methods and the importance to perform microarray screenings prior to qRT-PCR analyses were identified to be contributors to a lack of comparability. Also, pre-analytical variations like sample storage, degree of hemolysis, extraction efficiency and standardization methods are mentioned as important contributors to a reduced comparability[123]. The authors furthermore define the need for more large-scale studies with well-defined control- and validation cohorts limiting the influence of different HF etiologies, concomitant diseases, and treatments. Another important factor to be considered when interpreting miRNA quantification results is the influence of confounding medications and classical cardiovascular risk factors[123].
Taken together the very recent results in miRNA-based diagnostics of HFpEF and HFrEF are highly promising but urgently need verification in large-scale studies with harmonized methods and well-defined study samples.
Table 2 gives an overview of miRNAs in the diagnosis of HFpEF and HFrEF.
| miRNA | Study type | Bio-material | Groups/size | Detection method | Effect | Value as biomarker | Ref. |
| miR-211 | Experimental | Tissue | Mice - HFpEF vs HC | qRT-PCR | Up-regulated in HFpEF | Diagnostic | [67] |
| miR-30c | Clinical | Serum | HFpEF n = 90 | qRT-PCR | AUC analyses | Differentiating | [122] |
| HFrEF n = 90 | HFrEF from HFpEF | ||||||
| HC n = 90 | |||||||
| miR-125a-5p | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Up-regulated in HFrEF, | Diagnostic in HFrEF + | [115] |
| HFrEF n = 30 | unchanged in HFpEF | differentiating HFrEF | |||||
| HC n = 28 | from HFpEF | ||||||
| miR-146a | Clinical | Serum | HFpEF n = 90 | qRT-PCR | AUC analyses | Differentiating HFrEF | [122] |
| HFrEF n = 90 | from HFpEF | ||||||
| HC n = 90 | |||||||
| miR-183-3p1 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in | Diagnostic | [115] |
| HFrEF n = 30 | HFrEF and HFpEF | ||||||
| HC n = 28 | |||||||
| miR-190a* | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in | Diagnostic in HFpEF + | [115] |
| HFrEF n = 30 | HFpEF, unchanged in | differentiating | |||||
| HC n = 28 | HFrEF | HFrEF from HFpEF | |||||
| miR-193b-3p1 | clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in | Diagnostic | [115] |
| HFrEF n = 30 | HFrEF and HFpEF | ||||||
| HC n = 28 | |||||||
| miR-193b-5p1 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in HFpEF | Diagnostic | [115] |
| HFrEF n = 30 | |||||||
| HC n = 28 | |||||||
| miR-211-5p1 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in HFrEF | Diagnostic | [115] |
| HFrEF n = 30 | |||||||
| HC n = 28 | |||||||
| miR-221 | Clinical | Serum | HFpEF n = 90 | qRT-PCR | AUC analyses | Differentiating | [122] |
| HFrEF n = 90 | HFrEF from HFpEF | ||||||
| HC n = 90 | |||||||
| miR-328 | Clinical | Serum | HFpEF n = 90 | qRT-PCR | AUC analyses | Differentiating | [122] |
| HFrEF n = 90 | HFrEF from HFpEF | ||||||
| HC n = 90 | |||||||
| miR-375 | Clinical | Serum | HFpEF n = 90 | qRT-PCR | AUC analyses | Differentiating | [122] |
| HFrEF n = 90 | HFrEF from HFpEF | ||||||
| HC n = 90 | |||||||
| miR-454 | Clinical | Plasma | HFpEF n = 8 | qRT-PCR | Down-regulated in HFpEF | Diagnostic | [121] |
| Stable DCM n = 10 | |||||||
| Decompensated DCM n = 13 | |||||||
| HC n = 8 | |||||||
| miR-4941 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in HFrEF | Diagnostic | [115] |
| HFrEF n = 30 | |||||||
| HC n = 28 | |||||||
| miR-500 | Clinical | Plasma | HFpEF n = 8 | qRT-PCR | Down-regulated in HFpEF | Diagnostic | [121] |
| Stable DCM n = 10 | |||||||
| Decompensated DCM n = 13 | |||||||
| HC n = 8 | |||||||
| miR-545-5p | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Up-regulated in HFpEF | Diagnostic | [115] |
| HFrEF n = 30 | |||||||
| HC n = 28 | |||||||
| miR-550a-5p | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Up-regulated in HFrEF | Differentiating | [115] |
| HFrEF n = 30 | Down-regulation in HFpEF | HFrEF from HFpEF | |||||
| miR-638 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Down-regulated in HFrEF, | Diagnostic in HFrEF + | [115] |
| HFrEF n = 30 | unchanged in HFpEF | differentiating HFrEF | |||||
| HC n = 28 | from HFpEF | ||||||
| miR-671-5p1 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Up-regulated in HFrEF | Diagnostic | [115] |
| HFrEF n = 30 | |||||||
| HC n = 28 | |||||||
| miR-12331 | Clinical | Plasma | HFpEF n = 30 | qRT-PCR | Up-regulated in HFpEF | Diagnostic | [115] |
| HFrEF n = 30 | |||||||
| HC n = 28 | |||||||
| miR-1246 | Clinical | Plasma | HFpEF n = 8 | qRT-PCR | Down-regulated in HFpEF | Diagnostic | [121] |
| Stable DCM n = 10 | |||||||
| Decompensated DCM n = 13 | |||||||
| HC n = 8 |
Molecular diagnostics and therapeutics represent an important contributor to improve outcome for HF[123]. In contrast to traditional treatments, gene therapy is capable of modifying the genetic structure of the cell and can modulate the disease phenotype[124]. In this respect miRNAs are promising new players in the development of molecular therapeutics in cardiovascular disease and HF in particular. The regulation of selected miRNAs highly involved in cardiac remodeling could be a key factor in influencing the development of HF by controlling hypertrophy and fibrosis.
The concept of miRNA related disease treatment bases on the idea to specifically influence miRNA levels by raising or suppressing miRNA levels. Several different approaches have successfully been tested[125]. The major method to raise miRNA levels is the miRNA mimic technology (miR-mimic), which operates via miRNA substitution by artificially generated double-stranded miRNA-like RNA fragments[126]. They “mimic” endogenous miRNAs and bind - unlike endogenous miRNAs - gene-specifically to their target mRNA[126]. On the other hand, so called antagomirs can be used to suppress miRNA levels[127]. Antagomirs are chemically engineered oligonucleotides that competitively bind to and thus inhibit the mature target miRNA[127,128]. This mechanism leads to an up-regulation of specific mRNAs and gene expression[129]. Furthermore, miRNA sponges (also referred to as “target mimicry”) are competitive inhibitors that contain binding sites for a miRNA family and thus inactivate miRNAs of that particular family[130-132]. As opposed to antagomirs, sponges are specific only to the seed region of a miRNA and thus can interfere with a whole family of miRNAs[133]. Masking (also called “target occupiers”) describes a mechanism to prevent specific miRNAs from binding to their very binding site[134]. Consequently, fewer miRNAs remain to bind to the target and their effect is lessened. Erasers are oligonucleotides complementary to a specific miRNA. By binding to the miRNA the eraser inhibits its endogenous function[127].
Up to now there are no clinical trials published dealing with miRNA therapeutics in humans. Therefore, the following results relate to in-vitro studies and animal models. Initial studies in the field of miRNA therapeutics in HF were designed to identify differential regulations of miRNAs in HF. Sucharov et al[135] extracted miRNAs from 6 nonfailing, 5 idiopathic dilated cardiomyopathy (IDC) and 5 ischemic dilated cardiomyopathy (ISC) patients. The authors were able to find a set of miRNAs dysregulated in both IDC and ISC. In order to further evaluate the function of increased or decreased expression of those miRNAs the group introduced virus-delivered mirmimics as well as antagomirs against miR-92, miR-100 and miR-133b into neonatal rat ventricular myocytes and was able to cause dramatic down-regulation or up-regulation of the particular miRNAs[135]. Pathophysiologically, an up-regulation of miR-100 resulted in repression of adult genes αMyHC and SERCA while fetal genes ANF and βMyHC were up-regulated. These observations suggest the involvement of miR-100 in the specific regulation of gene expression involved in the repression of adult isoforms. The study shows that an artificial dysregulation of miR-100 is able to affect HF associated gene expression.
An initial project studying miRNA-associated therapeutic aspects in HF with respect to specifically raising miRNA levels was performed by Karakikes et al[136]. The authors addressed previous findings that proved miR-1 to be a key regulator of cardiac hypertrophy[44-47,90,137] and analyzed whether the restauration of miR-1 expression has protective effects on maladaptive cardiac remodeling. They established a hypertrophy and ventricular dilatation model in rats by ascending aortic banding before they raised miR-1 expression in-vivo by systemically administered adeno-associated virus-mediated gene transfer[136]. The authors were able to detect improved systolic as well as diastolic LV function in the miR-1 restauration group as measured by echocardiography and catheter-based pressure-volume loop analyses[136].
A similar approach was pursued by Pan et al[138] who induced an adenovirus-mediated overexpression of miR-101a in rats with chronic MI and were able to find a significant improvement of cardiac performance in those subjects treated with miR-101a overexpression. Recently, these results were confirmed in a rat model of induced MI[139]. The authors found decreased miR-101a levels at the site of the infarction and were able to verify this observation in cultured cardiac fibroblasts exposed to hypoxia and linked this effect to a TGF-β-modulated fibrotic effect. An administration of miR-101a mirmimics reduced the expression of TGF-β[139] indicating that miR-101a mimicry might negatively regulate fibrosis in ischemic cardiac tissue. These findings point out a potential applicability of mirmimics in the field of HF therapy and ignited studies further evaluating this aspect. In a recent study the authors succeeded in modulating myocardial fibrosis and apoptosis in a hypertrophic mouse model by regulating miR-455 levels[140]. Tail vein injection of viral delivered miR-455 resulted in aggravated cardiac hypertrophy on the one hand but also reduced myocardial fibrosis and inhibited apoptosis suggesting that this treatment can prevent maladaptive ventricular remodeling[140].
A different approach was addressed by Dakhlallah et al[141]. The authors used mirmimics to raise miR-133a levels in mesenchymal stem cells (MSC) and implanted these into ischemic rat hearts. Compared to non-miR-133a treated MSCs these rat hearts were shown to have increased cardiac function, decreased fibrosis and presented with improved cell engraftment due to better survival of miR-133a treated MSCs[141]. These study results indicate a potential roll of miRNAs in HF treatment with respect to an improvement of bioengineering of stem cells and are an example of the broad potential applicability of miRNAs in the field of HF treatment.
Reduced miR-29 levels were observed to be associated with a decrease of cardiac remodeling in mice[142]. The authors used an ischemia/reperfusion model in mice to analyze the effect of miR-29 on post-infarction remodeling. They found that an antisense inhibition of miR-29 implemented by an antagomir against miR-29 inhibited post infarction/reperfusion apoptosis and necrosis and led to a reduction of cardiac remodeling[142]. In a rat model of aortic constriction-induced HFpEF revealing higher cellular miR-21 levels in HFpEF rats compared to sham operated animals[67] (see above), Dong et al[67] performed further analyses after administering a miR-21 antagonist. The authors were able to find a reduction of fibrosis in those rats’ cardiac tissues that were transfected with anti-miR-21 and attributed this effect to a reduction of Bcl-2 expression - an anti-apoptotic factor involved in the apoptosis of cardiac fibroblasts[67]. The same miRNA was analyzed in a transgenic mouse model of cardiac failure. Thum et al[55] were able to show that in-vivo silencing of miR-21 by a systemically applied specific antagomir inhibits interstitial fibrosis by a reduction of mitogen-activated protein kinase activity when applied to pressure-overload-induced cardiac dysfunction in mice.
In order to assess the therapeutic potential of miR-652 in another mouse model with established pathological hypertrophy and cardiac dysfunction due to induced pressure overload Bernardo et al[143] first proved miR-652 expression to be elevated in pressure overloaded hearts compared to healthy controls. The authors then systemically administered antimiR-652 and found the expression of miR-652 effectively silenced in heart tissue of treated mice. The authors were able to show that antimiR-652 treated mice had better cardiac function and improved cardiac diameters compared to controls[143].
The first discovered miRNA Let-7 recently was found to be a potential therapeutic target in the treatment of deteriorated cardiac function after MI[144]. After induced MI in mice Let-7 was inhibited with a specific systemically applied antagomir. Molecularly, the expression of pluripotency-associated genes Oct4 and Sox2 was increased in cardiac fibroblasts in vitro and in vivo. Let-7 antagomir treated mice showed preserved LVEF and improved cardiac output compared to controls[144].
Addressing the hallmark of phathological hypertrophy and HF - the reactivation of fetal cardiac genes, in which miR-208 is highly involved, study results were reported in a model of antagomir-based silencing of miR-208. Montgomery et al[145] were able to silence miR-208 in a rat model of diastolic HF (Dahl salt-sensitive rats) by means of systemically administered locked nucleic acid-modified antimirs. On the one hand the authors found pathological myosin switching and cardiac remodeling lessened in antagomir-208 treated animals. More important from a clinical point of view was the observation that in diastolic HF therapeutic silencing of miR-208 resulted in lessened HF symptoms, a reduction of cardiac remodeling and an improved cardiac function as well as longer survival compared to control animals[145].
Current treatment strategies in HF are predominantly focused on HFrEF and no distinct therapy is established with respect to HFpEF[146-149]. The current non-specific therapy of HFpEF is limited and requires development and improvement of more distinct diagnostic and therapeutic options. Molecular diagnostics and therapeutics might provide the foundation for differential therapeutic approaches with regards to HFpEF and HFrEF.
Numerous studies have proven miRNAs to be key regulators and moderators in the development of HF and its pathophysiological precursors hypertrophy and fibrosis. Their molecular construction and integration in cellular and intercellular transport mechanisms define miRNAs as ideal circulating biomarkers for diagnostic and prognostic purposes while they can easily be collected and analyzed. Therefore, the application of miRNAs as circulating biomarkers represents a promising tool to complement established protein-based biomarkers of HF such as NPs on the one hand or novel stand-alone biomarkers in the diagnosis and prognosis of HF. In the differential diagnostics of HFrEF and HFpEF miRNAs can reliably differentiate between these two disease entities, although this has to be confirmed in larger samples. This is especially interesting considering the fact that the diagnosis of HFpEF at an early stage might significantly improve secondary prevention and established biomarkers of HF still lack precision in the differentiated diagnosis of HFpEF. Nevertheless, looking at the large number of studies only few of them confirmed previous findings with identical results and still different miRNAs are identified to be linked to HF presumably reflecting the complex interaction of miRNAs and their target sites. In this respect analysis of combinations of several miRNAs - miRNA signatures - represent a promising way to increase diagnostic and prognostic accuracy. An important aspect that should get attention when performing miRNA analyses is the comparability and standardization of analytical methods and the need for well-defined study samples.
Over the past years several different possibilities have been identified to alter levels of circulating miRNAs by systemically administering agents such as mirmimics or antagomirs. This therapeutic approach has been reported to significantly reduce hypertrophy and CF and improve LV function in animal models. It represents a promising approach to complement existing therapeutic options in the treatment of HF. Nevertheless, results of in-vitro and in-vivo models have not yet led to an application in clinical studies. A successful implementation of those insights in clinical trials represents the next step towards realizing this idea.
P- Reviewer: Iacoviello M, Rassaf T, Teragawa H S- Editor: Gong ZM L- Editor: A E- Editor: Liu SQ
| 1. | Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3501] [Cited by in F6Publishing: 3443] [Article Influence: 149.7] [Reference Citation Analysis (0)] |
| 2. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8672] [Cited by in F6Publishing: 8409] [Article Influence: 271.3] [Reference Citation Analysis (0)] |
| 3. | Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80:193-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 366] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 4. | Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF, Jeyaseelan K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81:617-627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 5. | Fan HM, Sun XY, Guo W, Zhong AF, Niu W, Zhao L, Dai YH, Guo ZM, Zhang LY, Lu J. Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. J Psychiatr Res. 2014;59:45-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 6. | Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2287] [Cited by in F6Publishing: 2246] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 7. | Kalozoumi G, Yacoub M, Sanoudou D. MicroRNAs in heart failure: Small molecules with major impact. Glob Cardiol Sci Pract. 2014;2014:79-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T, Müller-Ardogan M. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 904] [Cited by in F6Publishing: 920] [Article Influence: 65.7] [Reference Citation Analysis (0)] |
| 9. | Gläser C, Wichmann T, Wagner U, Gabert A, Schneider R. [Scanning electron microscopy studies in the detection of apo-B,E receptor activity of the lymphocyte membrane for diagnostic verification of genetically determined disorders of lipid metabolism]. Wien Klin Wochenschr. 1988;100:613-618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 440] [Cited by in F6Publishing: 448] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 10. | Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499-506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 563] [Cited by in F6Publishing: 578] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 11. | Gomes da Silva AM, Silbiger VN. miRNAs as biomarkers of atrial fibrillation. Biomarkers. 2014;19:631-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc Diagn Ther. 2015;5:17-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 57] [Reference Citation Analysis (0)] |
| 13. | Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2408] [Cited by in F6Publishing: 2352] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 14. | Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616-1626. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1577] [Cited by in F6Publishing: 1424] [Article Influence: 64.7] [Reference Citation Analysis (0)] |
| 15. | Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M. A uniform system for microRNA annotation. RNA. 2003;9:277-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1324] [Cited by in F6Publishing: 1279] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 16. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26776] [Article Influence: 1338.8] [Reference Citation Analysis (0)] |
| 17. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2105] [Cited by in F6Publishing: 2246] [Article Influence: 132.1] [Reference Citation Analysis (0)] |
| 18. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5833] [Cited by in F6Publishing: 6223] [Article Influence: 388.9] [Reference Citation Analysis (0)] |
| 19. | van Rooij E. The art of microRNA research. Circ Res. 2011;108:219-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 387] [Cited by in F6Publishing: 392] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 20. | Bauersachs J, Thum T. Biogenesis and regulation of cardiovascular microRNAs. Circ Res. 2011;109:334-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483-495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 722] [Cited by in F6Publishing: 771] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 22. | Dzikiewicz-Krawczyk A. MicroRNA polymorphisms as markers of risk, prognosis and treatment response in hematological malignancies. Crit Rev Oncol Hematol. 2015;93:1-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 24. | Salzman DW, Weidhaas JB. SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther. 2013;137:55-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 25. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5636] [Cited by in F6Publishing: 6046] [Article Influence: 377.9] [Reference Citation Analysis (0)] |
| 26. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1810] [Cited by in F6Publishing: 1937] [Article Influence: 138.4] [Reference Citation Analysis (0)] |
| 27. | Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 359] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 28. | Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e21-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1193] [Cited by in F6Publishing: 1502] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 29. | Levy D, Kenchaiah S, Larson MG, Benjamin EJ, Kupka MJ, Ho KK, Murabito JM, Vasan RS. Long-term trends in the incidence of and survival with heart failure. N Engl J Med. 2002;347:1397-1402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1514] [Cited by in F6Publishing: 1472] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 30. | McMurray JJ, Adamopoulos S, Anker SD, Auricchio A, Böhm M, Dickstein K, Falk V, Filippatos G, Fonseca C, Gomez-Sanchez MA. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association (HFA) of the ESC. Eur Heart J. 2012;33:1787-1847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3411] [Cited by in F6Publishing: 3448] [Article Influence: 287.3] [Reference Citation Analysis (0)] |
| 31. | Paulus WJ, Tschöpe C, Sanderson JE, Rusconi C, Flachskampf FA, Rademakers FE, Marino P, Smiseth OA, De Keulenaer G, Leite-Moreira AF. How to diagnose diastolic heart failure: a consensus statement on the diagnosis of heart failure with normal left ventricular ejection fraction by the Heart Failure and Echocardiography Associations of the European Society of Cardiology. Eur Heart J. 2007;28:2539-2550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1826] [Cited by in F6Publishing: 1800] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 32. | Farmakis D, Parissis J, Lekakis J, Filippatos G. Acute heart failure: Epidemiology, risk factors, and prevention. Rev Esp Cardiol (Engl Ed). 2015;68:245-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Dorn GW, Robbins J, Sugden PH. Phenotyping hypertrophy: eschew obfuscation. Circ Res. 2003;92:1171-1175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 196] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 34. | Chien KR. Stress pathways and heart failure. Cell. 1999;98:555-558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 342] [Cited by in F6Publishing: 337] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 35. | Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1103] [Cited by in F6Publishing: 1068] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 36. | van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255-18260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1230] [Cited by in F6Publishing: 1159] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 37. | Berk BC, Fujiwara K, Lehoux S. ECM remodeling in hypertensive heart disease. J Clin Invest. 2007;117:568-575. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 644] [Cited by in F6Publishing: 662] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 38. | Westermann D, Kasner M, Steendijk P, Spillmann F, Riad A, Weitmann K, Hoffmann W, Poller W, Pauschinger M, Schultheiss HP. Role of left ventricular stiffness in heart failure with normal ejection fraction. Circulation. 2008;117:2051-2060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 330] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 39. | Hrynchyshyn N, Jourdain P, Desnos M, Diebold B, Funck F. Galectin-3: a new biomarker for the diagnosis, analysis and prognosis of acute and chronic heart failure. Arch Cardiovasc Dis. 2013;106:541-546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 40. | Meluzín J, Tomandl J. Can biomarkers help to diagnose early heart failure with preserved ejection fraction? Dis Markers. 2015;2015:426045. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 41. | Holland DJ, Sacre JW, Leano RL, Marwick TH, Sharman JE. Contribution of abnormal central blood pressure to left ventricular filling pressure during exercise in patients with heart failure and preserved ejection fraction. J Hypertens. 2011;29:1422-1430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 42. | Borlaug BA, Nishimura RA, Sorajja P, Lam CS, Redfield MM. Exercise hemodynamics enhance diagnosis of early heart failure with preserved ejection fraction. Circ Heart Fail. 2010;3:588-595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 723] [Cited by in F6Publishing: 768] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 43. | Bauersachs J. Regulation of myocardial fibrosis by MicroRNAs. J Cardiovasc Pharmacol. 2010;56:454-459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 589] [Cited by in F6Publishing: 579] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 45. | Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193-2204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 301] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 46. | Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613-618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1343] [Cited by in F6Publishing: 1320] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 47. | Li Q, Song XW, Zou J, Wang GK, Kremneva E, Li XQ, Zhu N, Sun T, Lappalainen P, Yuan WJ. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci. 2010;123:2444-2452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 48. | Liu W, Liu Y, Zhang Y, Zhu X, Zhang R, Guan L, Tang Q, Jiang H, Huang C, Huang H. MicroRNA-150 Protects Against Pressure Overload-Induced Cardiac Hypertrophy. J Cell Biochem. 2015;116:2166-2176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137-1141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 343] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 50. | Song L, Su M, Wang S, Zou Y, Wang X, Wang Y, Cui H, Zhao P, Hui R, Wang J. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18:2266-2274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 96] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 51. | Kim JO, Song DW, Kwon EJ, Hong SE, Song HK, Min CK, Kim do H. miR-185 plays an anti-hypertrophic role in the heart via multiple targets in the calcium-signaling pathways. PLoS One. 2015;10:e0122509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 52. | Wang YS, Zhou J, Hong K, Cheng XS, Li YG. MicroRNA-223 displays a protective role against cardiomyocyte hypertrophy by targeting cardiac troponin I-interacting kinase. Cell Physiol Biochem. 2015;35:1546-1556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 53. | Hua Y, Zhang Y, Ren J. IGF-1 deficiency resists cardiac hypertrophy and myocardial contractile dysfunction: role of microRNA-1 and microRNA-133a. J Cell Mol Med. 2012;16:83-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 54. | Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831-1840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 387] [Cited by in F6Publishing: 412] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 55. | Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980-984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1770] [Cited by in F6Publishing: 1822] [Article Influence: 113.9] [Reference Citation Analysis (0)] |
| 56. | Busk PK, Cirera S. MicroRNA profiling in early hypertrophic growth of the left ventricle in rats. Biochem Biophys Res Commun. 2010;396:989-993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 57. | van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1265] [Cited by in F6Publishing: 1221] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 58. | Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772-2786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 642] [Cited by in F6Publishing: 623] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 59. | Shyu KG, Wang BW, Wu GJ, Lin CM, Chang H. Mechanical stretch via transforming growth factor-β1 activates microRNA208a to regulate endoglin expression in cultured rat cardiac myoblasts. Eur J Heart Fail. 2013;15:36-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 60. | Hirt MN, Werner T, Indenbirken D, Alawi M, Demin P, Kunze AC, Stenzig J, Starbatty J, Hansen A, Fiedler J. Deciphering the microRNA signature of pathological cardiac hypertrophy by engineered heart tissue- and sequencing-technology. J Mol Cell Cardiol. 2015;81:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 61. | da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567-1576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 239] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 62. | Wang BW, Wu GJ, Cheng WP, Shyu KG. MicroRNA-208a increases myocardial fibrosis via endoglin in volume overloading heart. PLoS One. 2014;9:e84188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 63. | Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106:166-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 290] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 64. | Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465-472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 260] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 65. | Castaldi A, Zaglia T, Di Mauro V, Carullo P, Viggiani G, Borile G, Di Stefano B, Schiattarella GG, Gualazzi MG, Elia L. MicroRNA-133 modulates the β1-adrenergic receptor transduction cascade. Circ Res. 2014;115:273-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 66. | Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170-18, 6p following 178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 644] [Cited by in F6Publishing: 693] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 67. | Dong S, Ma W, Hao B, Hu F, Yan L, Yan X, Wang Y, Chen Z, Wang Z. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int J Clin Exp Pathol. 2014;7:565-574. [PubMed] [Cited in This Article: ] |
| 68. | Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res. 2009;82:21-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 459] [Cited by in F6Publishing: 478] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 69. | Cardin S, Guasch E, Luo X, Naud P, Le Quang K, Shi Y, Tardif JC, Comtois P, Nattel S. Role for MicroRNA-21 in atrial profibrillatory fibrotic remodeling associated with experimental postinfarction heart failure. Circ Arrhythm Electrophysiol. 2012;5:1027-1035. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 70. | Liang H, Zhang C, Ban T, Liu Y, Mei L, Piao X, Zhao D, Lu Y, Chu W, Yang B. A novel reciprocal loop between microRNA-21 and TGFβRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol. 2012;44:2152-2160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 71. | Villar AV, García R, Merino D, Llano M, Cobo M, Montalvo C, Martín-Durán R, Hurlé MA, Nistal JF. Myocardial and circulating levels of microRNA-21 reflect left ventricular fibrosis in aortic stenosis patients. Int J Cardiol. 2013;167:2875-2881. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 72. | Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 295] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 73. | Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, Chun B, Zhuang J, Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 264] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 74. | Wang J, Huang W, Xu R, Nie Y, Cao X, Meng J, Xu X, Hu S, Zheng Z. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J Cell Mol Med. 2012;16:2150-2160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 212] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 75. | Ramdas V, McBride M, Denby L, Baker AH. Canonical transforming growth factor-β signaling regulates disintegrin metalloprotease expression in experimental renal fibrosis via miR-29. Am J Pathol. 2013;183:1885-1896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 76. | Nijhuis A, Biancheri P, Lewis A, Bishop CL, Giuffrida P, Chan C, Feakins R, Poulsom R, Di Sabatino A, Corazza GR. In Crohn’s disease fibrosis-reduced expression of the miR-29 family enhances collagen expression in intestinal fibroblasts. Clin Sci (Lond). 2014;127:341-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 77. | Zhu H, Fan GC. Role of microRNAs in the reperfused myocardium towards post-infarct remodelling. Cardiovasc Res. 2012;94:284-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 127] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 78. | Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590-11593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 463] [Cited by in F6Publishing: 496] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 79. | Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133-6140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 625] [Cited by in F6Publishing: 663] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 80. | Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 473] [Cited by in F6Publishing: 495] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 81. | Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369-381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 465] [Cited by in F6Publishing: 486] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 82. | van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027-13032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1319] [Cited by in F6Publishing: 1415] [Article Influence: 88.4] [Reference Citation Analysis (0)] |
| 83. | Yang F, Li P, Li H, Shi Q, Li S, Zhao L. microRNA-29b Mediates the Antifibrotic Effect of Tanshinone IIA in Postinfarct Cardiac Remodeling. J Cardiovasc Pharmacol. 2015;65:456-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 84. | Mann DL, Bristow MR. Mechanisms and models in heart failure: the biomechanical model and beyond. Circulation. 2005;111:2837-2849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 614] [Cited by in F6Publishing: 640] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 85. | Mann DL. MicroRNAs and the failing heart. N Engl J Med. 2007;356:2644-2645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 86. | Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 701] [Cited by in F6Publishing: 703] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 87. | Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res. 2008;103:1072-1083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 194] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 88. | Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111-2116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 434] [Cited by in F6Publishing: 435] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 89. | Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 90. | Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 472] [Cited by in F6Publishing: 451] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 91. | Leptidis S, El Azzouzi H, Lok SI, de Weger R, Olieslagers S, Kisters N, Silva GJ, Heymans S, Cuppen E, Berezikov E. A deep sequencing approach to uncover the miRNOME in the human heart. PLoS One. 2013;8:e57800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 92. | Satoh M, Minami Y, Takahashi Y, Tabuchi T, Nakamura M. Expression of microRNA-208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J Card Fail. 2010;16:404-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 93. | Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:186-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 303] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 94. | Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172-1187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1233] [Cited by in F6Publishing: 1300] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 95. | Iguchi H, Kosaka N, Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol. 2010;3:478-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 96. | Kuosmanen SM, Hartikainen J, Hippeläinen M, Kokki H, Levonen AL, Tavi P. MicroRNA profiling of pericardial fluid samples from patients with heart failure. PLoS One. 2015;10:e0119646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 97. | Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107:1047-1057. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 588] [Cited by in F6Publishing: 605] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 98. | Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 959] [Cited by in F6Publishing: 1005] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 99. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8246] [Cited by in F6Publishing: 9027] [Article Influence: 531.0] [Reference Citation Analysis (0)] |
| 100. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003-5008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2345] [Cited by in F6Publishing: 2488] [Article Influence: 191.4] [Reference Citation Analysis (0)] |
| 101. | Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1953] [Cited by in F6Publishing: 2062] [Article Influence: 158.6] [Reference Citation Analysis (0)] |
| 102. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3218] [Cited by in F6Publishing: 3383] [Article Influence: 211.4] [Reference Citation Analysis (0)] |
| 103. | Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402-4407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 891] [Cited by in F6Publishing: 910] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 104. | McManus DD, Ambros V. Circulating MicroRNAs in cardiovascular disease. Circulation. 2011;124:1908-1910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 105. | Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, Glaab WE. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977-1983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 477] [Cited by in F6Publishing: 473] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 106. | Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575-581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1601] [Cited by in F6Publishing: 1657] [Article Influence: 110.5] [Reference Citation Analysis (0)] |
| 107. | Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG, van Rooij E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15:650-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 108. | Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 488] [Cited by in F6Publishing: 495] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 109. | Kumarswamy R, Anker SD, Thum T. MicroRNAs as circulating biomarkers for heart failure: questions about MiR-423-5p. Circ Res. 2010;106:e8; author reply e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 110. | Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14:147-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 252] [Cited by in F6Publishing: 266] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 111. | Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357-2366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 390] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 112. | Topkara VK, Mann DL. Role of microRNAs in cardiac remodeling and heart failure. Cardiovasc Drugs Ther. 2011;25:171-182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 113. | Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138-1147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 114. | Marfella R, Di Filippo C, Potenza N, Sardu C, Rizzo MR, Siniscalchi M, Musacchio E, Barbieri M, Mauro C, Mosca N. Circulating microRNA changes in heart failure patients treated with cardiac resynchronization therapy: responders vs. non-responders. Eur J Heart Fail. 2013;15:1277-1288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 115. | Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, Chong JP, Ng JY, Chen YT, Chan MM. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015;17:393-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 116. | Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA. 2013;110:4255-4260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 274] [Cited by in F6Publishing: 272] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 117. | Gupta MK, Halley C, Duan ZH, Lappe J, Viterna J, Jana S, Augoff K, Mohan ML, Vasudevan NT, Na J. miRNA-548c: a specific signature in circulating PBMCs from dilated cardiomyopathy patients. J Mol Cell Cardiol. 2013;62:131-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 118. | Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet. 2011;4:614-619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 119. | Cakmak HA, Coskunpinar E, Ikitimur B, Barman HA, Karadag B, Tiryakioglu NO, Kahraman K, Vural VA. The prognostic value of circulating microRNAs in heart failure: preliminary results from a genome-wide expression study. J Cardiovasc Med (Hagerstown). 2015;16:431-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 120. | Vogel B, Keller A, Frese KS, Leidinger P, Sedaghat-Hamedani F, Kayvanpour E, Kloos W, Backe C, Thanaraj A, Brefort T. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J. 2013;34:2812-2822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 121. | Nair N, Kumar S, Gongora E, Gupta S. Circulating miRNA as novel markers for diastolic dysfunction. Mol Cell Biochem. 2013;376:33-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 122. | Watson CJ, Gupta SK, O’Connell E, Thum S, Glezeva N, Fendrich J, Gallagher J, Ledwidge M, Grote-Levi L, McDonald K. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. 2015;17:405-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 157] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 123. | Schmitter D, Voors AA, van der Harst P. HFpEF vs. HFrEF: can microRNAs advance the diagnosis? Eur J Heart Fail. 2015;17:351-354. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 124. | Katz MG, Fargnoli AS, Williams RD, Kendle AP, Steuerwald NM, Bridges CR. MiRNAs as potential molecular targets in heart failure. Future Cardiol. 2014;10:789-800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 125. | van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919-928. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 312] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 126. | Wang Z. The guideline of the design and validation of MiRNA mimics. Methods Mol Biol. 2011;676:211-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 127. | Caroli A, Cardillo MT, Galea R, Biasucci LM. Potential therapeutic role of microRNAs in ischemic heart disease. J Cardiol. 2013;61:315-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 128. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685-689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3005] [Cited by in F6Publishing: 2983] [Article Influence: 157.0] [Reference Citation Analysis (0)] |
| 129. | Oliveira-Carvalho V, Carvalho VO, Silva MM, Guimarães GV, Bocchi EA. MicroRNAs: a new paradigm in the treatment and diagnosis of heart failure? Arq Bras Cardiol. 2012;98:362-369. [PubMed] [Cited in This Article: ] |
| 130. | Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1634] [Cited by in F6Publishing: 1576] [Article Influence: 92.7] [Reference Citation Analysis (0)] |
| 131. | Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033-1037. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1408] [Cited by in F6Publishing: 1341] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 132. | Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858-R861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 371] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 133. | Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043-2050. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 523] [Cited by in F6Publishing: 543] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 134. | Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212:285-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 151] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 135. | Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. 2008;45:185-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 136. | Karakikes I, Chaanine AH, Kang S, Mukete BN, Jeong D, Zhang S, Hajjar RJ, Lebeche D. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc. 2013;2:e000078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 137. | Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377-2385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 298] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 138. | Pan Z, Sun X, Shan H, Wang N, Wang J, Ren J, Feng S, Xie L, Lu C, Yuan Y. MicroRNA-101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor-β1 pathway. Circulation. 2012;126:840-850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 246] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 139. | Zhao X, Wang K, Liao Y, Zeng Q, Li Y, Hu F, Liu Y, Meng K, Qian C, Zhang Q. MicroRNA-101a inhibits cardiac fibrosis induced by hypoxia via targeting TGFβRI on cardiac fibroblasts. Cell Physiol Biochem. 2015;35:213-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 140. | Wu C, Dong S, Li Y. Effects of miRNA-455 on cardiac hypertrophy induced by pressure overload. Int J Mol Med. 2015;35:893-900. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 141. | Dakhlallah D, Zhang J, Yu L, Marsh CB, Angelos MG, Khan M. MicroRNA-133a engineered mesenchymal stem cells augment cardiac function and cell survival in the infarct heart. J Cardiovasc Pharmacol. 2015;65:241-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 142. | Ye Y, Hu Z, Lin Y, Zhang C, Perez-Polo JR. Downregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2010;87:535-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 170] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 143. | Bernardo BC, Nguyen SS, Winbanks CE, Gao XM, Boey EJ, Tham YK, Kiriazis H, Ooi JY, Porrello ER, Igoor S. Therapeutic silencing of miR-652 restores heart function and attenuates adverse remodeling in a setting of established pathological hypertrophy. FASEB J. 2014;28:5097-5110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 144. | Tolonen AM, Magga J, Szabó Z, Viitala P, Gao E, Moilanen AM, Ohukainen P, Vainio L, Koch WJ, Kerkelä R. Inhibition of Let-7 microRNA attenuates myocardial remodeling and improves cardiac function postinfarction in mice. Pharmacol Res Perspect. 2014;2:e00056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 145. | Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537-1547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 459] [Cited by in F6Publishing: 443] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 146. | Nair N, Gupta S, Collier IX, Gongora E, Vijayaraghavan K. Can microRNAs emerge as biomarkers in distinguishing HFpEF versus HFrEF? Int J Cardiol. 2014;175:395-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 147. | Fonarow GC, Stough WG, Abraham WT, Albert NM, Gheorghiade M, Greenberg BH, O’Connor CM, Sun JL, Yancy CW, Young JB. Characteristics, treatments, and outcomes of patients with preserved systolic function hospitalized for heart failure: a report from the OPTIMIZE-HF Registry. J Am Coll Cardiol. 2007;50:768-777. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 743] [Cited by in F6Publishing: 801] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 148. | Gheorghiade M, Vaduganathan M, Fonarow GC, Bonow RO. Rehospitalization for heart failure: problems and perspectives. J Am Coll Cardiol. 2013;61:391-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 502] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 149. | Gheorghiade M, Shah AN, Vaduganathan M, Butler J, Bonow RO, Rosano GM, Taylor S, Kupfer S, Misselwitz F, Sharma A. Recognizing hospitalized heart failure as an entity and developing new therapies to improve outcomes: academics’, clinicians’, industry’s, regulators’, and payers’ perspectives. Heart Fail Clin. 2013;9:285-290, v-vi. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 150. | Fukushima Y, Nakanishi M, Nonogi H, Goto Y, Iwai N. Assessment of plasma miRNAs in congestive heart failure. Circ J. 2011;75:336-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |