Copyright
©The Author(s) 2016.
World J Biol Chem. Feb 26, 2016; 7(1): 188-205
Published online Feb 26, 2016. doi: 10.4331/wjbc.v7.i1.188
Published online Feb 26, 2016. doi: 10.4331/wjbc.v7.i1.188
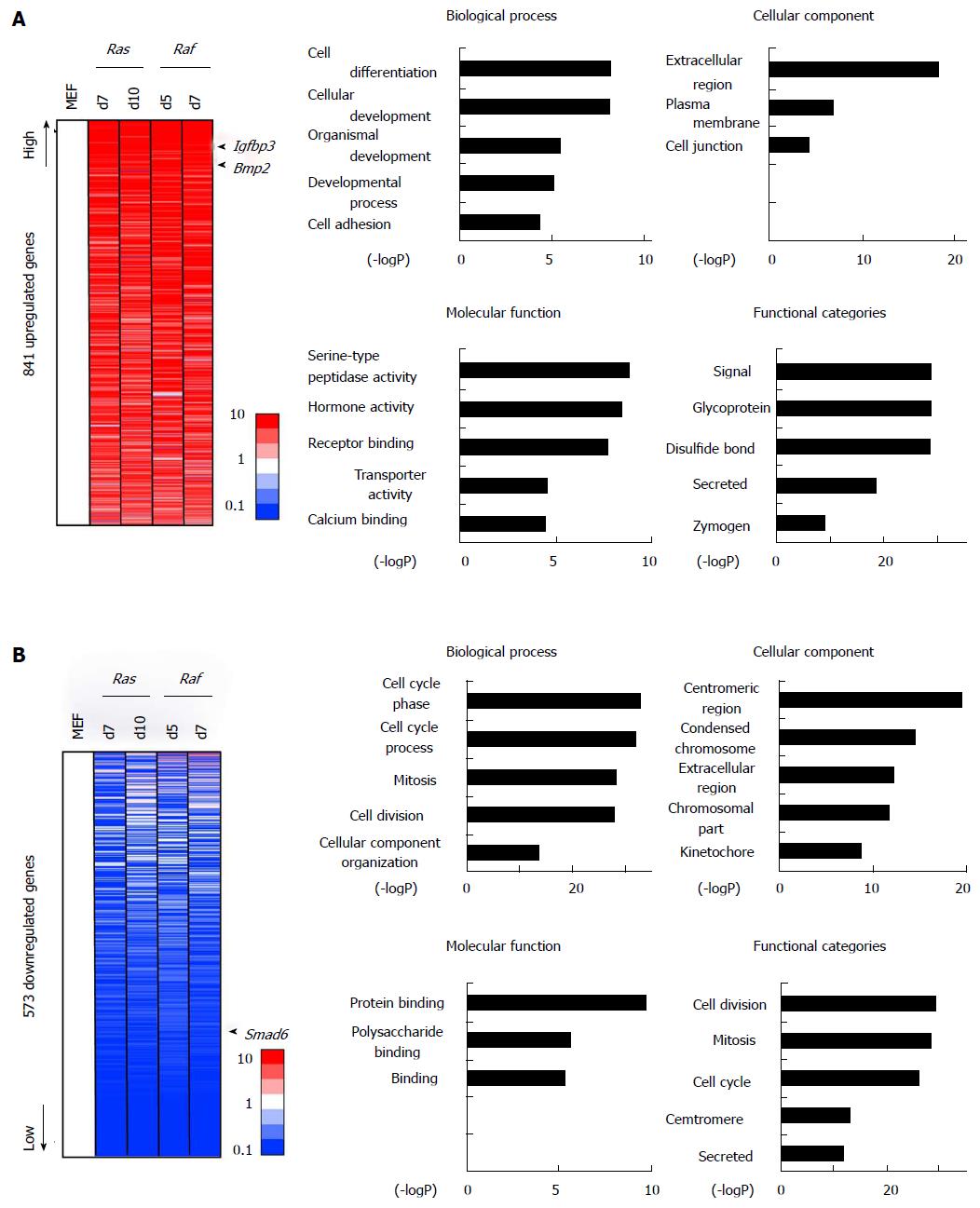
Figure 3 Gene annotation enrichment analysis for commonly altered genes.
A: Commonly upregulated genes. A total of 841 genes showed > 5-fold upregulation commonly in either of RasG12V cells on days 7 (Ras d7) and 10 (Ras d10) and in either of RafV600E cells on days 5 (Raf d5) and 7 (Raf d7), compared with MEF. More upregulated genes were sorted upward. Gene annotation enrichment was analyzed for biological process, cellular component, molecular function, and functional categories. Genes associated with signal/secreted proteins (P = 3.8 × 10-30), extracellular regions (P = 5.3 × 10-19), and cell differentiation/development (P = 6.1 × 10-9), e.g., Bmp2 and Igfbp3, were upregulated; B: Commonly downregulated genes. A total of 573 genes showed < 0.2-fold downregulation commonly in either of the two RasG12V cells and in either of the two RafV600E cells, compared with MEF. More downregulated genes were sorted downward. Gene annotation enrichment analysis showed that genes related to cell cycle (P = 1.3 × 10-33) such as Cdc6 were significantly enriched, in good agreement with growth arrest. Genes associated with secreted protein (P = 2.0 × 10-12) and extracellular region (P = 5.4 × 10-13) such as Cxcl5 and Wnt4 were also enriched, suggesting that the dynamic activation and repression of secreted factors occurred commonly during Ras- and Raf-induced senescence. The GO-term “protein binding” also showed significant enrichment, and Smad6 was included in this term. MEF: Mouse embryonic fibroblast.
- Citation: Fujimoto M, Mano Y, Anai M, Yamamoto S, Fukuyo M, Aburatani H, Kaneda A. Epigenetic alteration to activate Bmp2-Smad signaling in Raf-induced senescence. World J Biol Chem 2016; 7(1): 188-205
- URL: https://www.wjgnet.com/1949-8454/full/v7/i1/188.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v7.i1.188