Copyright
©2014 Baishideng Publishing Group Inc.
World J Biol Chem. May 26, 2014; 5(2): 180-203
Published online May 26, 2014. doi: 10.4331/wjbc.v5.i2.180
Published online May 26, 2014. doi: 10.4331/wjbc.v5.i2.180
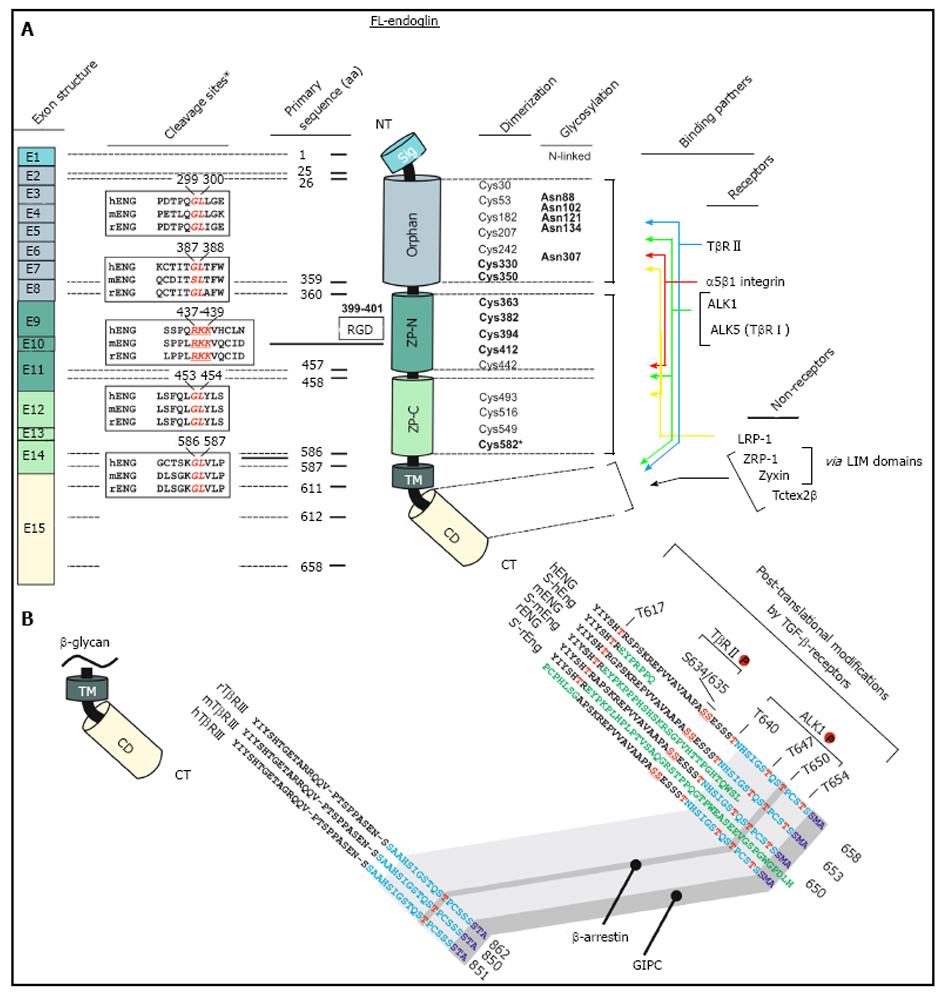
Figure 1 Schematic representation of the structural and functional modules of human endoglin.
A: Left, first panel, Exon structure: Structure of the endoglin gene and assignment of respective protein modules. Left, second panel, cleavage sites: Predicted consensus proteolytical cleavage sites deduced from the primary sequence and experimentally confirmed matrix metalloproteinases-14 cleavage site between aa positions 586 and 587[59]. Middle panel, primary sequence positions: aa boundaries of the structural domains of human endoglin and location of the Arginine-Glycin-Aspartic acid (RGD) sequence that is only present in human endoglin. Right, first panel, dimerization: The cysteine residues of the extracellular domain of endoglin are depicted including the 8 highly conserved residues within the ZP-domain[47]. Cysteines that are involved in dimerization are shown in bold. Cysteine 582 that is involved in human endoglin dimerization is not present in the mouse and rat homologues[56]. Right, second panel, glycosylation: This figure part displays verified N-linked glycosylation sites within human endoglin. There is experimental evidence for O-linked glycosylation, but respective sites are not shown[1]. Right, third panel, binding partners: Depicted are interaction partners of endoglin as either receptor proteins (upper) or cytosolic non receptors proteins (lower). Colored arrows indicate interacting domains of endoglin with its binding partners. Zyxin and ZRP-1 bind to endoglin via their LIM-domains; B: Displayed is an aa alignment of betaglycan (left) and endoglin (right) of human, mouse and rat. Receptor kinase substrates (serine and threonines) are shown in red. Threonine 650 is essential for binding to β-arrestin2[66]. The C-termini of betaglycan and endoglin that are highly conserved are indicated in light blue. The PDZ-I domain which binds to GIPC is depicted in dark blue[67]. The alternative C-termini which results from differential splicing are shown in green.
- Citation: Meurer SK, Alsamman M, Scholten D, Weiskirchen R. Endoglin in liver fibrogenesis: Bridging basic science and clinical practice. World J Biol Chem 2014; 5(2): 180-203
- URL: https://www.wjgnet.com/1949-8454/full/v5/i2/180.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v5.i2.180