Copyright
©The Author(s) 2016.
World J Biol Chem. Aug 26, 2016; 7(3): 223-230
Published online Aug 26, 2016. doi: 10.4331/wjbc.v7.i3.223
Published online Aug 26, 2016. doi: 10.4331/wjbc.v7.i3.223
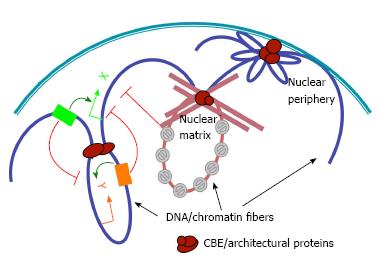
Figure 1 The roles of chromatin boundary elements in organizing genomic and nuclear architecture.
Chromatin boundary element (CBE) complexes (brown ovals) may interact with each other (left), nuclear matrix (middle) or nuclear envelope (right) to tether chromatin loops that modulate enhancer-promoter interactions (curved arrows), and block the spread of silent chromatin (grey forbidden circles). Green and orange arrows indicate promoters of hypothetical genes X and Y, respectively. Green and orange boxes, enhancer elements for genes X and Y, respectively.
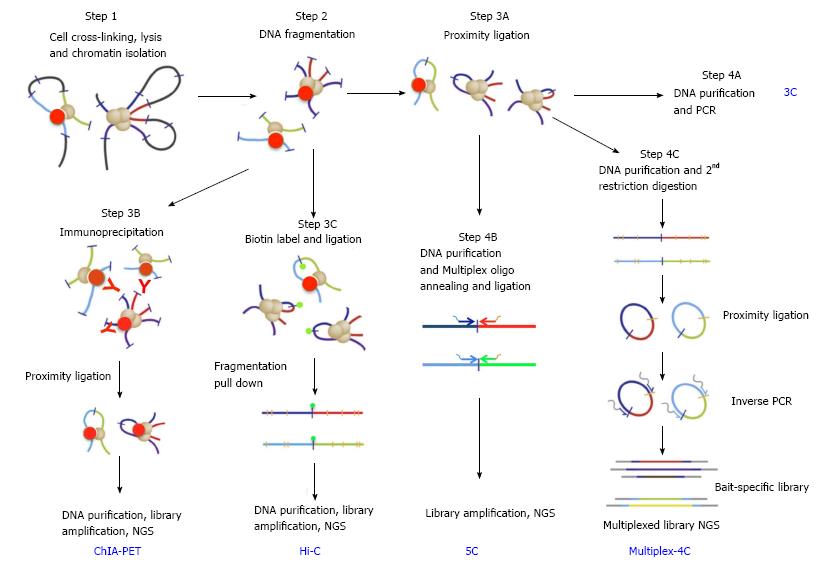
Figure 2 Key steps in chromosomal conformation capture protocols (adapted from Stadhouders et al[46], 2013).
Bold blue upper case letters denote capture protocols and black lower case phrases denote the key steps. Steps 1 and 2 (cell crosslinking, chromatin preparation and fragmentation) are common to all capture protocols. Following these, the ends of captured DNA fragments are ligated under highly diluted condition (step 3A, proximity ligation), and junctions of captured DNAs are detected by PCR using pray- and bait-specific primers (step 4A, 3C). Alternatively, capture events can be selected by immunoprecipitation using antibody against specific architectural proteins (step 3B for ChIA-PET). Capture events can be also enriched by pull-down of Biotin-labeled ligation junction (step 3C for Hi-C). After purification, libraries of junction DNAs are amplified and sequenced by next-generation-sequencing (NGS). After step 3A, library of bait- and pray-specific primers can be used in conjunction (step 4B for 5C) to generate libraries of selected capture junctions for NGS sequencing. Bait-specific primers can also be used to generate a capture library (step 4C for 4C or multiplex 4C).
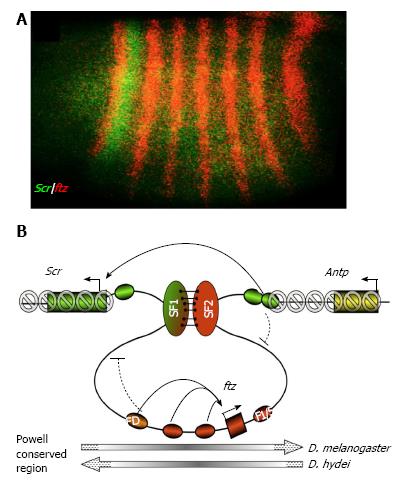
Figure 3 Hypothetical function of the SF1-SF2 chromatin loop in Scr and ftz gene regulation (adapted from Li et al[27], 2015).
A: Segmental expression patter of a Drosophila Hox gene Scr (green). A neighbor non-Hox gene ftz is expressed in pair-rule stripes (red); B: A model for developmentally regulated SF1-SF2 chromatin loop in modulating enhancer-promoter communications and preventing spread of silent chromatin in the Drosophila Hox complex. The chromatin loop anchored by the interaction between SF1 and SF2 (green and red vertical ovals, respectively) during early Drosophila embryogenesis can restrict the access of ftz and Scr enhancers (red and green ovals, respectively) to neighboring gene promoters (curved block signs). The SF1-SF2 loop also prevents the spreading of the repressive H3K9Me3 and H3K27Me3 histone modification (grey forbidden circles) into the active ftz gene. Pairing of SF1 and SF2 may facilitate “enhancer bypass”, connect the Scr with its distal enhancers (green ovals) in 3D in the early Drosophila embryo. The ftz gene domain is present in opposite orientation in various Drosophila species (grey long arrows, bottom). FD: Ftz distal enhancer; F1/5: Ftz stripe 1/5 enhancer; Antp: Antennapedia, a Hox gene located distal to ftz.
- Citation: Ma Z, Li M, Roy S, Liu KJ, Romine ML, Lane DC, Patel SK, Cai HN. Chromatin boundary elements organize genomic architecture and developmental gene regulation in Drosophila Hox clusters. World J Biol Chem 2016; 7(3): 223-230
- URL: https://www.wjgnet.com/1949-8454/full/v7/i3/223.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v7.i3.223