Copyright
©2011 Baishideng Publishing Group Co.
World J Biol Chem. May 26, 2011; 2(5): 90-97
Published online May 26, 2011. doi: 10.4331/wjbc.v2.i5.90
Published online May 26, 2011. doi: 10.4331/wjbc.v2.i5.90
Figure 1 Multiple templates approach sequence alignment.
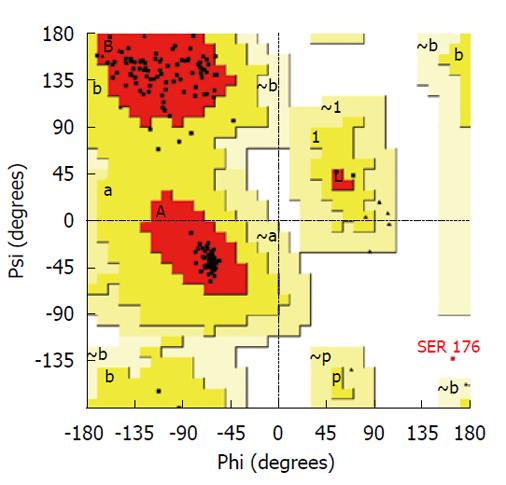
Figure 2 Ramachandran plot of model resulting from loop refinement of multiple template model.
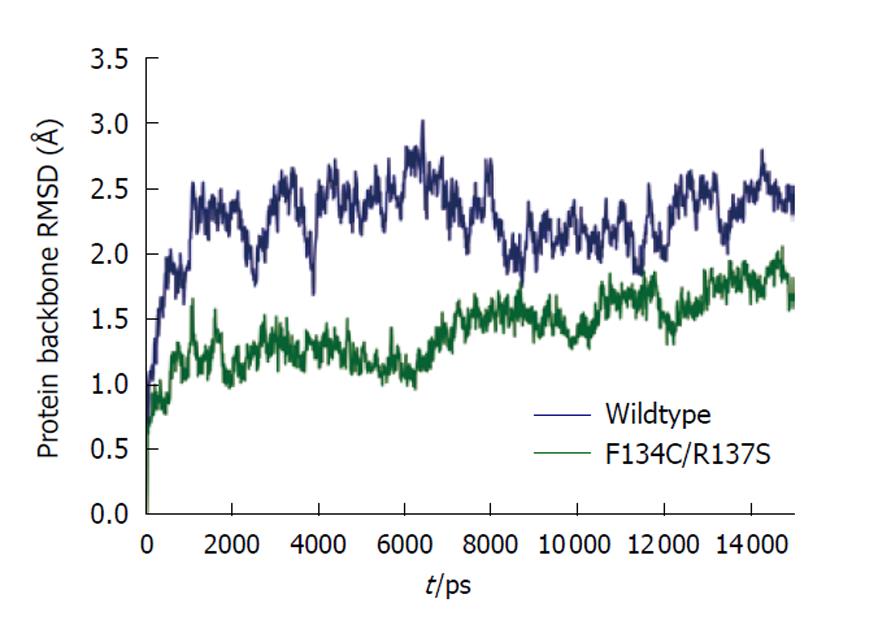
Figure 3 Molecular dynamics protein backbone atom root mean square deviation plot.
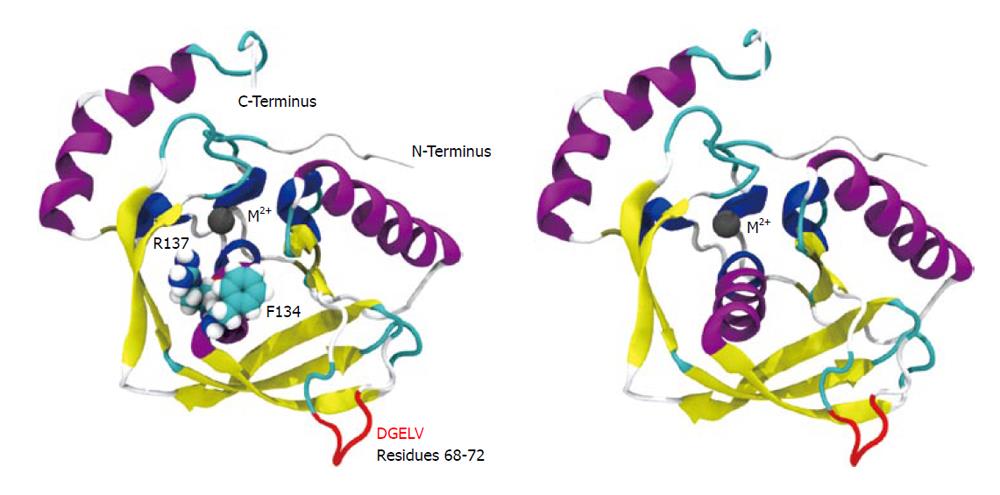
Figure 4 Cartoon depiction of wild-type structure showing Phe134 and Arg137 near the active site (left) and the “clean” overall structure model (right).
Figure 5 Increased atomic motion in loop residues Asp68 through Val72 (DGELV) in the F134C/R137S chlamydial peptide deformylase as reflected by average per residue backbone atom temperature factor.
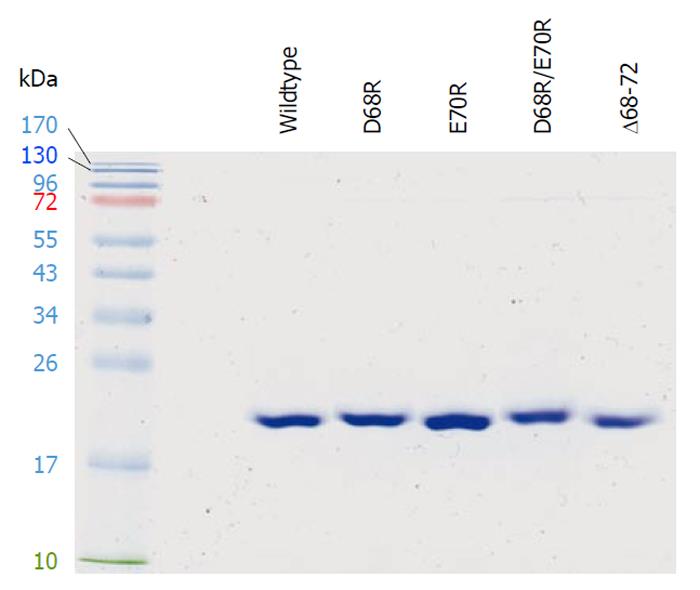
Figure 6 Purified recombinant chlamydial peptide deformylase proteins.
Chlamydial peptide deformylases carrying a (His)6-tag were expressed in Escherichia coli, purified by cobalt agarose, resolved by 13.5% SDS-polyacrylamide gel, and visualized by Coomassie blue staining. In the left lane are protein standards with molecular weights indicated.
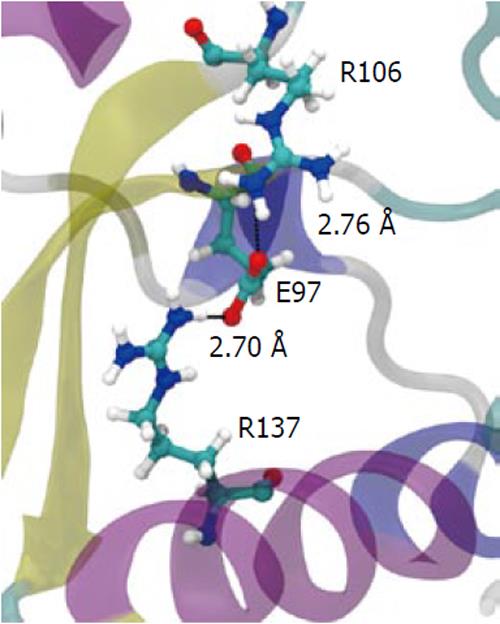
Figure 7 The salt-bridge “sandwich” in the wild-type.
Residues Arg137, Glu97 and Arg106 form a tight salt bridge adding rigidity to the structure in the active site region. When broken, Arg106 is pulled more tightly into the active site.
Figure 8 Least squares fit of wild-type with F134C/R137S mutant backbone atoms.
The wild-type is colored blue and the mutant red. Note shift of residue range Glu63 to Glu67 in the large loop region by 0.29 nm towards the active site. The overall average RMSD fit for the protein backbone atoms was 0.15 nm.
- Citation: Oey CB, Bao X, Lewis C, Kerrigan JE, Fan H. High tolerance to mutations in a Chlamydia trachomatis peptide deformylase loop. World J Biol Chem 2011; 2(5): 90-97
- URL: https://www.wjgnet.com/1949-8454/full/v2/i5/90.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v2.i5.90